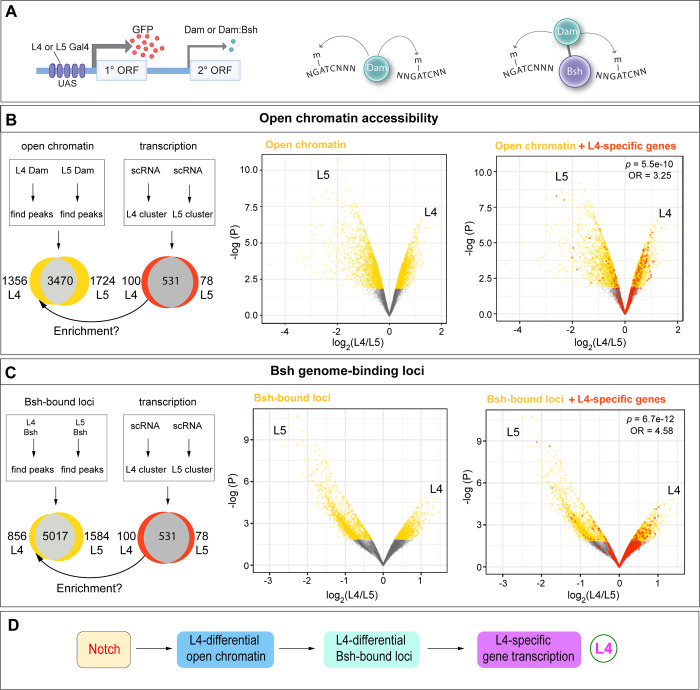
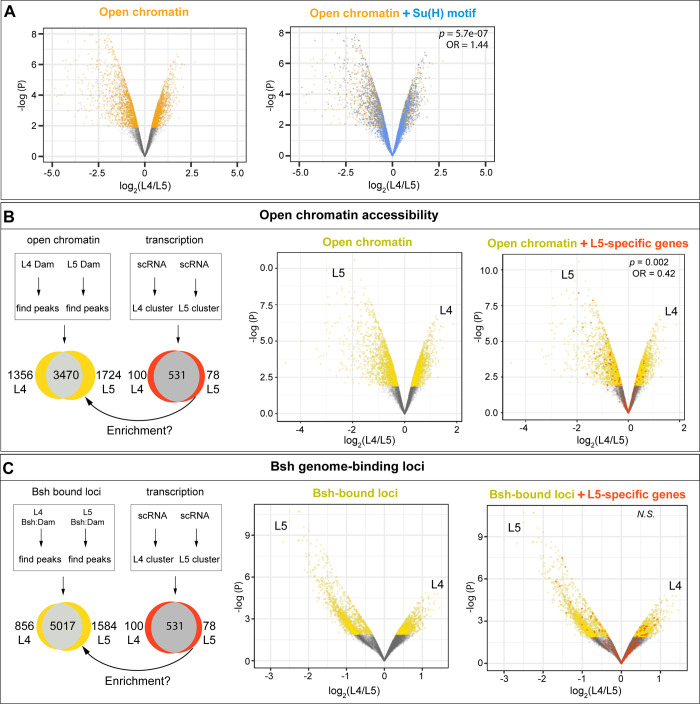
Figure 6. The NotchON L4 has correlated open chromatin, Bsh-bound loci, and transcription profile that is distinct from the NotchOFF L5.
(A) Schematic of the Targeted DamID method. Upon GAL4 induction, low levels of either Dam or Bsh:Dam are expressed, allowing genome-wide open chromatin and Bsh binding targets to be identified. (B) L4 and L5 neurons show distinct open chromatin regions (yellow), and L4-specific transcribed genes (red) are enriched in L4-distinct open chromatin regions. (C) L4 and L5 neurons show distinct Bsh-bound loci (yellow), and L4-specific transcribed genes (red) are enriched in L4-distinct Bsh-bound loci. p Values from Fisher’s exact test; odds ratios (OR) expressed as L4/L5 in all cases. (D) Model.