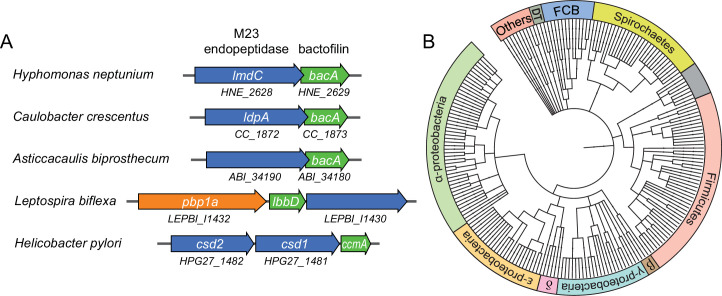
Figure 7. Putative operons of including adjacent BacA and M23 peptidase genes are widely conserved among bacteria.
(A) Arrangement of bacA and lmdC genes in species whose BacA homolog has a proven role in cell morphogenesis. (B) Co-conservation of BacA and M23 peptidase genes across different bacterial phyla. Bioinformatic analysis was used to identify genomes that contained a bactofilin gene located next to an M23 peptidase gene. After retrieval of the taxonomy IDs of the corresponding species from the National Center for Biotechnology Information (NCBI) website, a phylogenetic tree of the species was created using the iTOL server (Letunic and Bork, 2021). Abbreviations: DT (Deinococcus-Thermus group), FCB (Fibrobacteres-Chlorobi-Bacteroidetes group), β (β-proteobacteria), δ (δ-proteobacteria). The full list of species used and details on the genes identified are provided in Figure 7—source data 1.