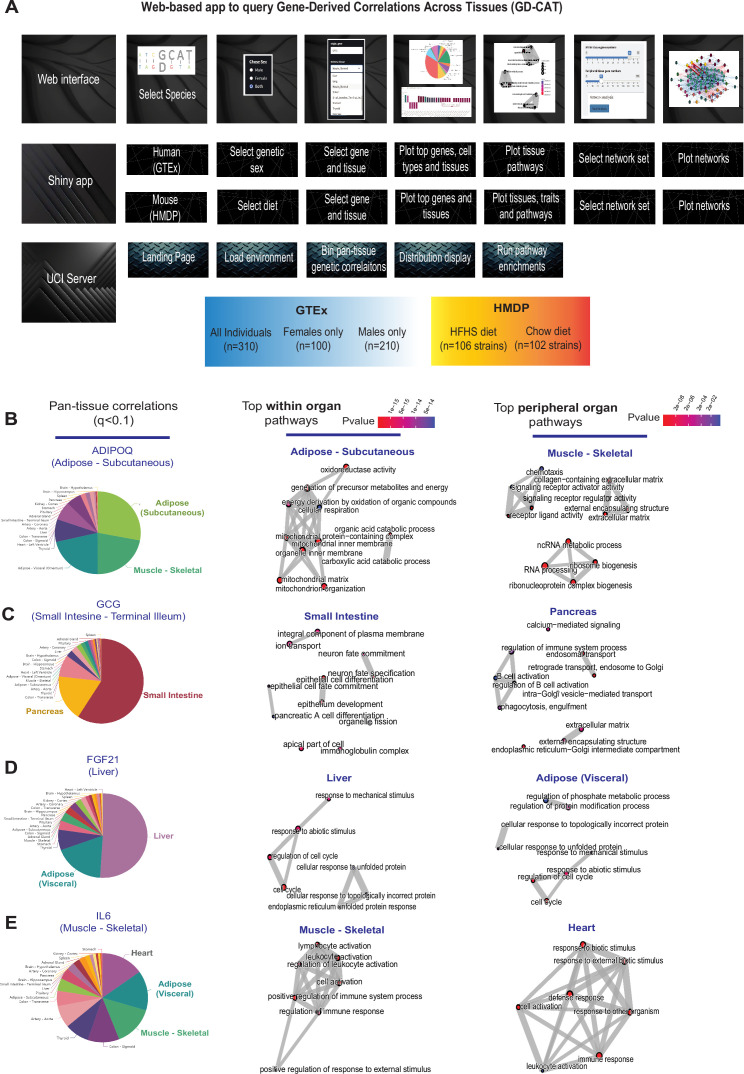
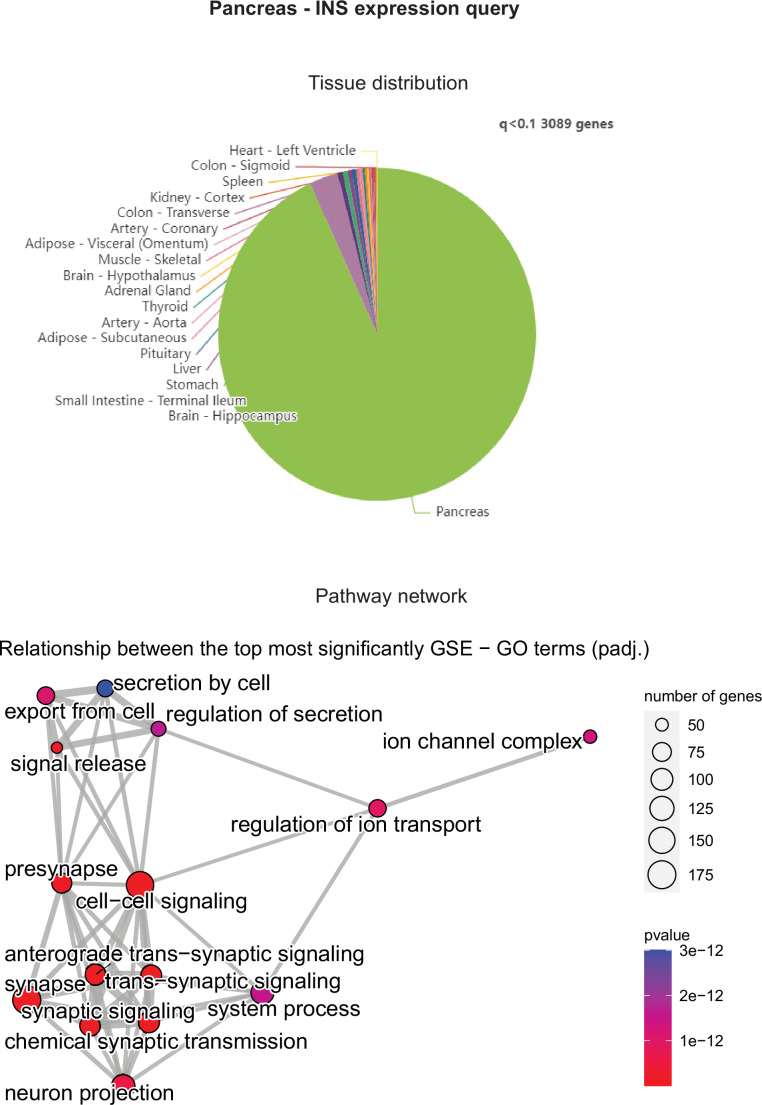
Figure 1. Web tool overview and inter-individual correlation structure of established endocrine proteins.
(A) Web server structure for user-defined interactions, as well as server and shiny app implementation scheme for gene-derived correlations across tissues (GD-CAT). (B) All genes across the 18 metabolic tissues in 310 individuals were correlated with expression of ADIPOQ in subcutaneous adipose tissue, where a q-value cutoff of q<0.1 showed the strongest enrichments with subcutaneous and muscle gene expression (pie chart, left). Gene set enrichment analysis (GSEA) was performed using the bicor coefficient of all genes to ADIPOQ using gene ontology biological process annotations and network construction of top pathways using clusterprofiler, where pathways related to fatty acid oxidation were observed in adipose (left) and chemotaxis/ECM remodeling in skeletal muscle (right). (B–D) The same q-value binning, top within-tissue and top peripheral enrichments were applied to intestinal GCG (C), liver FGF21 (D), and muscle IL6 (E). For these analyses all 310 individuals (across both sexes) were used and q-value adjustments calculated using a Benjamini-Hochberg FDR adjustment.