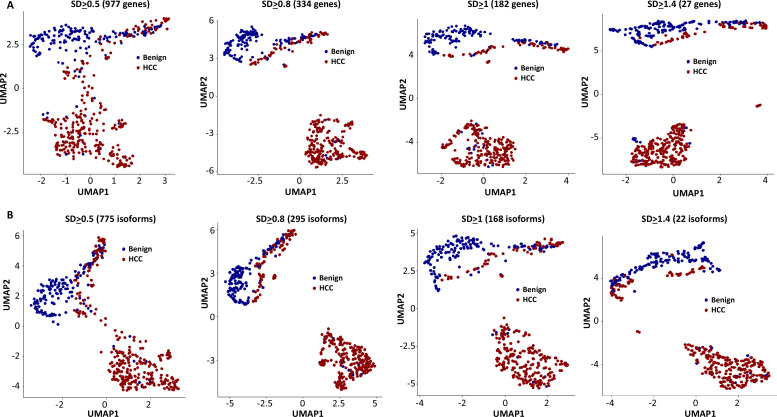
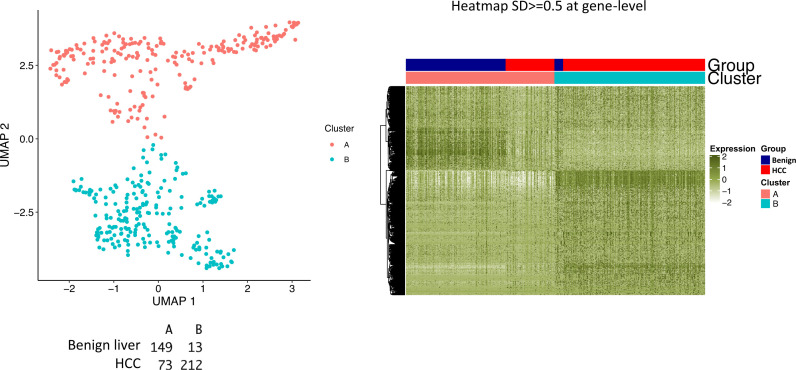
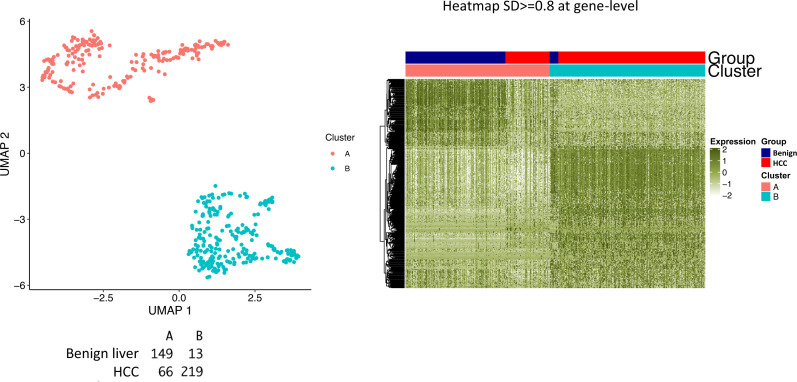
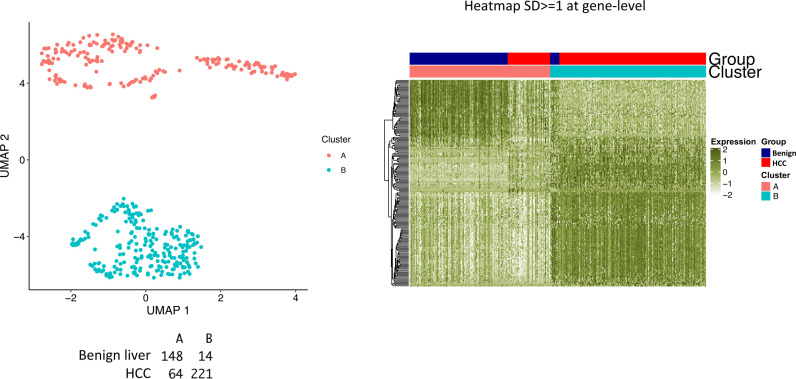
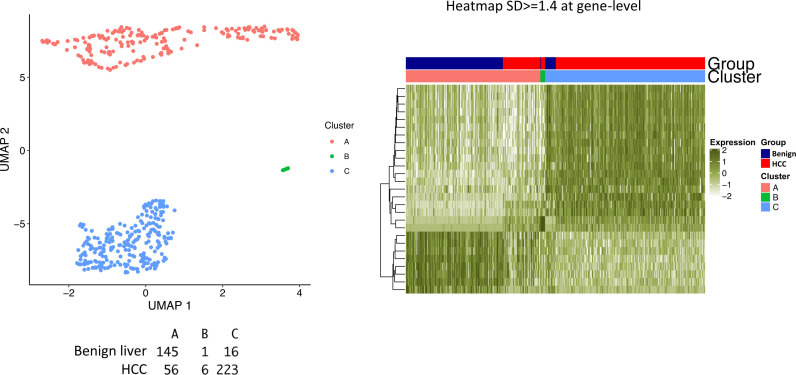
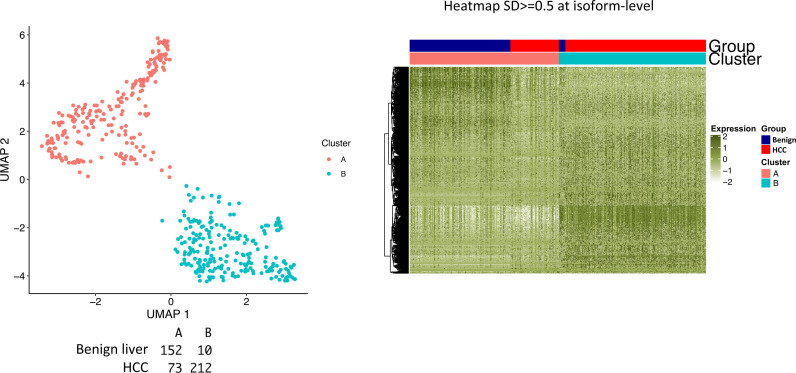
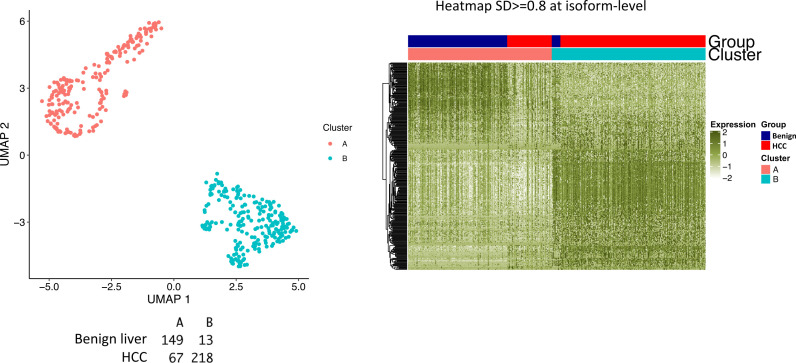
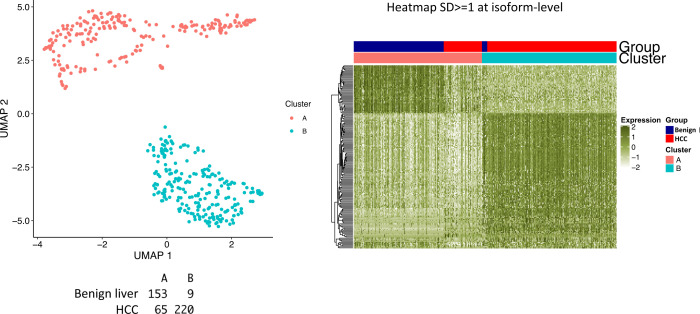
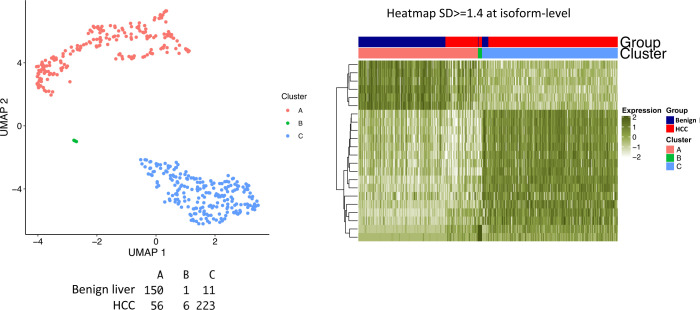
Figure 12. Uniform Manifold Approximation and Projection (UMAP) cluster analyses of cells from the hepatocellular carcinoma (HCC) and benign liver based on gene expressions with different standard deviation cutoffs.
(A) UMAP clustering of cells based on gene expression with standard deviations at least 0.5, 0.8, 1.0, or 1.4. The numbers of genes employed in the UMAP analysis are indicated. Blue dot, cell from the benign liver; red dot, cell from HCC. (B) UMAP clustering of cells based on isoform expressions with standard deviations at least 0.5, 0.8, 1.0, or 1.4. The numbers of genes employed in the UMAP analysis are indicated. Blue dot, cell from the benign liver; red dot, cell from HCC.