Figure 3. Single-nucleus gene expression in the human locus coeruleus (LC) using single-nucleus RNA-sequencing (snRNA-seq).
We applied an unsupervised clustering workflow to identify cell populations in the snRNA-seq data. (A) Unsupervised clustering identified 30 clusters representing populations including norepinephrine (NE) neurons (red), 5-HT neurons (purple), and other major neuronal and non-neuronal cell populations (additional colors). Marker genes (columns) were used to identify clusters (rows). Cluster IDs are shown in labels on the right, and the numbers of nuclei per cluster are shown in horizontal bars on the right. Percentages of nuclei per cluster are also shown in Figure 3—figure supplement 1D. Heatmap values represent mean logcounts per cluster. (B) UMAP representation of nuclei, with colors matching cell populations from heatmap. (C) Differential expression (DE) testing between neuronal clusters identified a total of 327 statistically significant genes with elevated expression in the NE neuron cluster, at a false discovery rate (FDR) threshold of 0.05 and fold-change (FC) threshold of 2. Heatmap displays the top 70 genes, ranked in descending order by FDR, excluding mitochondrial genes, with NE neuron marker genes described in text highlighted in red. The full list of 327 genes including mitochondrial genes is provided in Supplementary file 2C. Heatmap values represent mean logcounts in the NE neuron cluster and mean logcounts per cluster averaged across all other neuronal clusters (excluding ambiguous). (D–E) Cross-species comparison showing expression of human ortholog genes for LC-associated genes identified in the rodent LC (Mulvey et al., 2018; Grimm et al., 2004) using alternative experimental technologies. Boxplots show logcounts per nucleus in the NE neuron cluster and all other neuronal clusters. Boxplot whiskers extend to 1.5 times the interquartile range, and outliers are not shown. (F) DE testing between neuronal clusters identified a total of 361 statistically significant genes with elevated expression in the 5-HT neuron cluster, at an FDR threshold of 0.05 and FC threshold of 2. Heatmap displays the top 70 genes, ranked in descending order by FDR, with 5-HT neuron marker genes described in text highlighted in red. The full list of 361 genes is provided in Supplementary file 2F.
Figure 3—figure supplement 1. Distribution of nucleus-level quality control (QC) metrics across unsupervised clusters in snRNA-seq data.
Figure 3—figure supplement 2. Additional quality control (QC) evaluations for NE neuron cluster in snRNA-seq data.
Figure 3—figure supplement 3. Supervised identification of NE neuron nuclei by thresholding on expression of NE neuron marker genes in snRNA-seq data.
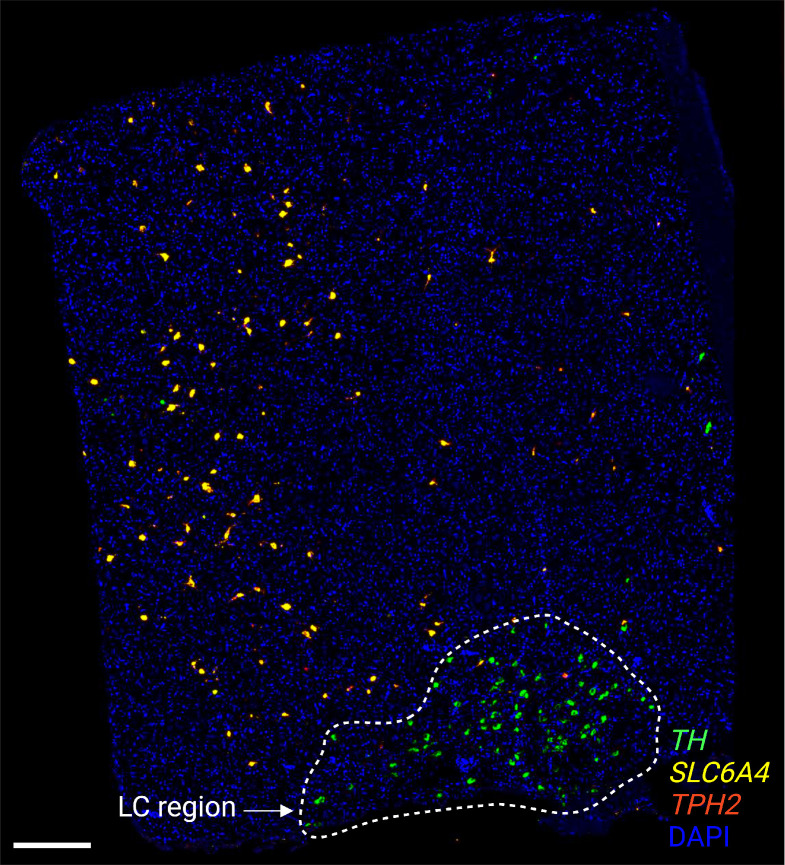
Figure 3—figure supplement 4. Expression of NE neuron marker genes in individual cells using RNAscope and high-magnification confocal imaging.
Figure 3—figure supplement 5. DE testing results between neuronal clusters in the LC and surrounding region in snRNA-seq data.
Figure 3—figure supplement 6. DE testing results between NE neuron cluster and all other clusters in snRNA-seq data.
Figure 3—figure supplement 7. Overlap and comparison between DE genes identified in SRT and snRNA-seq data.
Figure 3—figure supplement 8. Unsupervised clustering results showing additional inhibitory neuronal, miscellaneous, dopaminergic, and cholinergic marker genes in snRNA-seq data.
Figure 3—figure supplement 9. Spatial expression and enrichment analysis of 5-HT neuron marker genes in Visium SRT samples.
Figure 3—figure supplement 10. Spatial expression of additional marker genes for 5-HT neurons in Visium SRT samples.