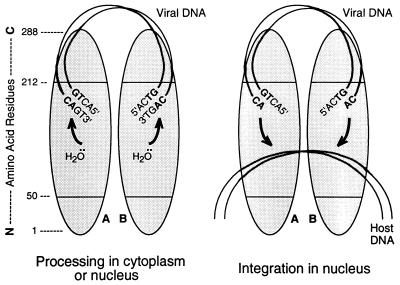
FIG. 8.
Model for the functional organization of the IN protein. Protein monomers A and B (shaded ovals) are shown as a dimer, in parallel orientations, with a linear map of amino acid positions at the extreme left, numbered from the N terminus to the C terminus. Initially (left), viral DNA is bound nonspecifically by the C-terminal region of IN. Amino acids in the central region (perhaps residues 191 to 212) recognize the terminal six positions of the viral DNA sequence (only the final four positions are shown) and position the CA bases for attack by a water molecule that is positioned by the C-terminal portion of the active site (defined by the three invariant acidic amino acids in the central region). The terminal 3′-OH of the viral DNA itself can be used instead of water to release cyclic (GT) dinucleotides (21). In the nucleus (right), host DNA is bound by other portions of the central region of IN. Following a conformational change in the protein, each processed viral DNA end is positioned by the C-terminal portion of the active site, with the aid of the residues responsible for viral DNA recognition and perhaps the C-terminal region, to attack the host DNA. During nonspecific alcoholysis, small alcohols can be used by the active site without requiring residues outside of numbers 50 to 190. The model would accommodate actions occurring in trans or involving multimers larger than dimers.