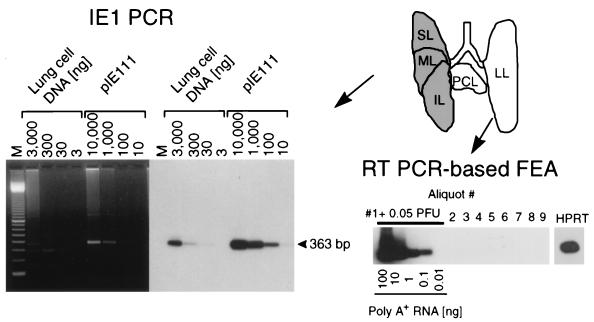
FIG. 3.
Verification of CMV latency in the lungs. The analysis is shown for the group with no CD8 T-cell therapy. (Top right) Scheme of the lobular anatomy of the lungs in ventral view is depicted at the upper right. For each individual mouse included in a latency analysis performed 12 months after infection, the left lung (LL) and postcaval lobe (PCL) were used to verify the absence of infectious virus by the RT-PCR-based focus expansion assay (FEA) whereas the superior lobe (SL), middle lobe (ML), and inferior lobe (IL) were used to detect the presence of latent viral DNA. (Bottom right) Nine 2-ml aliquots of the homogenate of the LL and PCL were tested for the presence of infectivity in cultures of permissive cells by the RT-PCR-based focus expansion assay. As a positive control, aliquot 1 was supplemented with 0.05 PFU of purified murine CMV. Poly(A)+ RNA derived from this culture was serially diluted as indicated, whereas for each of the remaining cultures, 100 ng of poly(A)+ RNA was subjected to ie1 exon 3/4-specific RT-PCR, leading to an amplification product of 188 bp. The autoradiograph was obtained after hybridization with a γ-32P-end-labeled oligonucleotide probe directed against the splice junction. For culture 9, the presence of RNA was verified by an RT-PCR specific for the HPRT housekeeping gene transcript. (Left) DNA isolated from the SL, ML, and IL was subjected to an ie1 exon 4-specific PCR. Plasmid pIE111 was added to pulmonary DNA from uninfected BMT recipients (10,000 copies in 3 μg) and was titrated in parallel. Amplification products were analyzed by gel electrophoresis. Lane M contains the 100-bp size marker kit. An ethidium bromide-stained gel is shown on the left, and the corresponding Southern blot autoradiograph obtained after hybridization with a γ-32P-end-labeled internal oligonucleotide probe is shown on the right.