FIGURE 9.
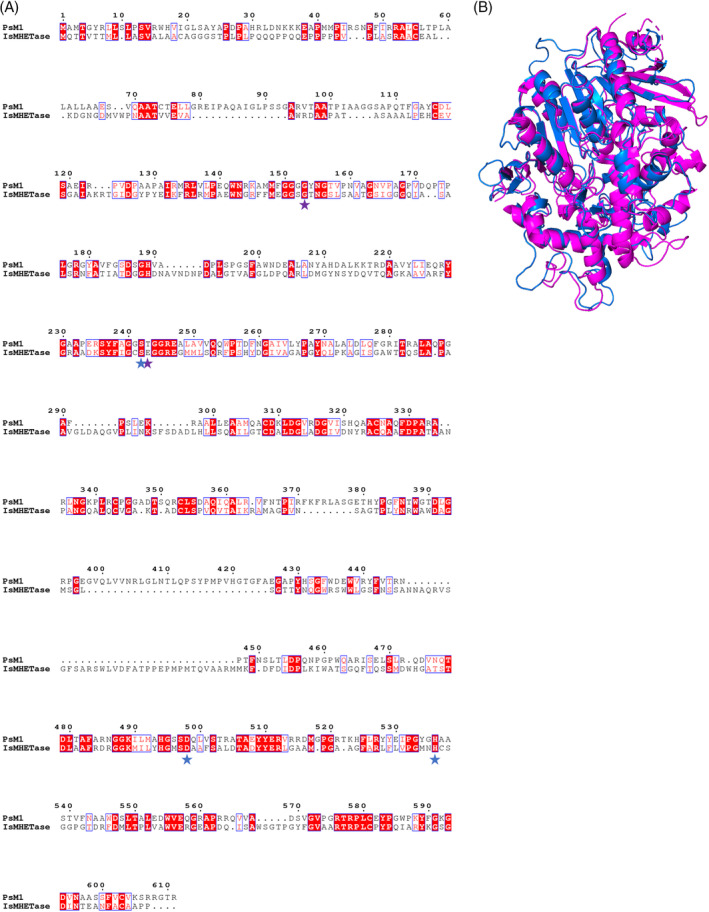
Protein sequence structural alignment of PsM1 and IsMHETase. (A) Generated using T‐COFFEE in Expresso mode and rendered in ESPript 3.0. Amino acids shaded in red represent identical residues, while those outlined in blue share similar biochemical properties. Catalytic triad residues are marked with blue stars and oxyanion hole residues are marked by purple stars. (B) PyMOL structural alignment of Phyre2 predicted PsM1 (blue) and IsMHETase (PDB 6QGB) (magenta) with a RMSD of 3.631573 over 504 residues, it is noted that IsMHETase structure starts on residue 43, which may increase the RMSD value.
