Figure 8.
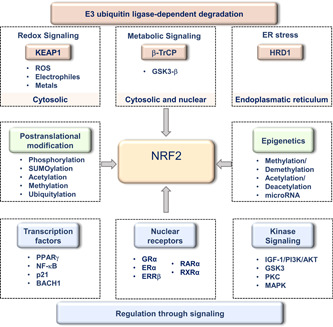
NRF2‐regulatory pathways. The transcription factor NRF2 consists of multiple domains that are under tight regulatory control. In complexes with KEAP1, NRF2 is ubiquitinated and degraded by the proteasome. Electrophilic and oxidative modifications of cysteine residues in KEAP1 lower the affinity to NRF2, and factors like p21 or NF‐κB hamper the binding of KEAP1 to NRF2. Ubiquitin‐dependent NRF2 degradation is further determined through the availability of the substrate recognition component of the SKP1‐cullin 1‐F‐box protein E3 ligase complex β‐TrCP, which responds to metabolic changes and is regulated by GSK3‐β. Another NRF2‐regulatory factor is the E3 ubiquitin‐protein ligase HRD1 which participates in ER‐associated degradation during ER stress. A variety of signaling cascades regulate the expression, nuclear availability, DNA‐binding affinity, and transactivation activity of NRF2, and also posttranslational and epigenetic modifications essentially contribute to the regulation of NRF2 activity. These pathways and factors represent potential targets for NRF2‐inhibiting small molecules. AKT, protein kinase B; BACH1, BTB domain and CNC homolog 1; ERRβ, estrogen‐related receptor β; ERα, estrogen receptor α; GRα, glucocorticoid receptor α; GSK3‐β, glycogen synthase kinase‐3β; HRD1, HMG‐CoA reductase degradation protein 1; IGF‐1, insulin‐like growth factor 1; MAPK, mitogen‐activated protein kinase; NF‐κB, nuclear factor κ‐light‐chain‐enhancer of activated B cells; NRF2, nuclear factor erythroid 2‐related factor 2; PI3K, phosphatidylinositol 3‐kinase; PKC, protein kinase C, PPARγ, peroxisome proliferator‐activated receptor γ; RARα, retinoic acid receptor α; RXRα, retinoid X receptor α. [Color figure can be viewed at wileyonlinelibrary.com]
