Fig. 5. G allele of KMT2E SNV rs73184087 binds HIF-2α to control the KMT2E-AS1/KMT2E pair.
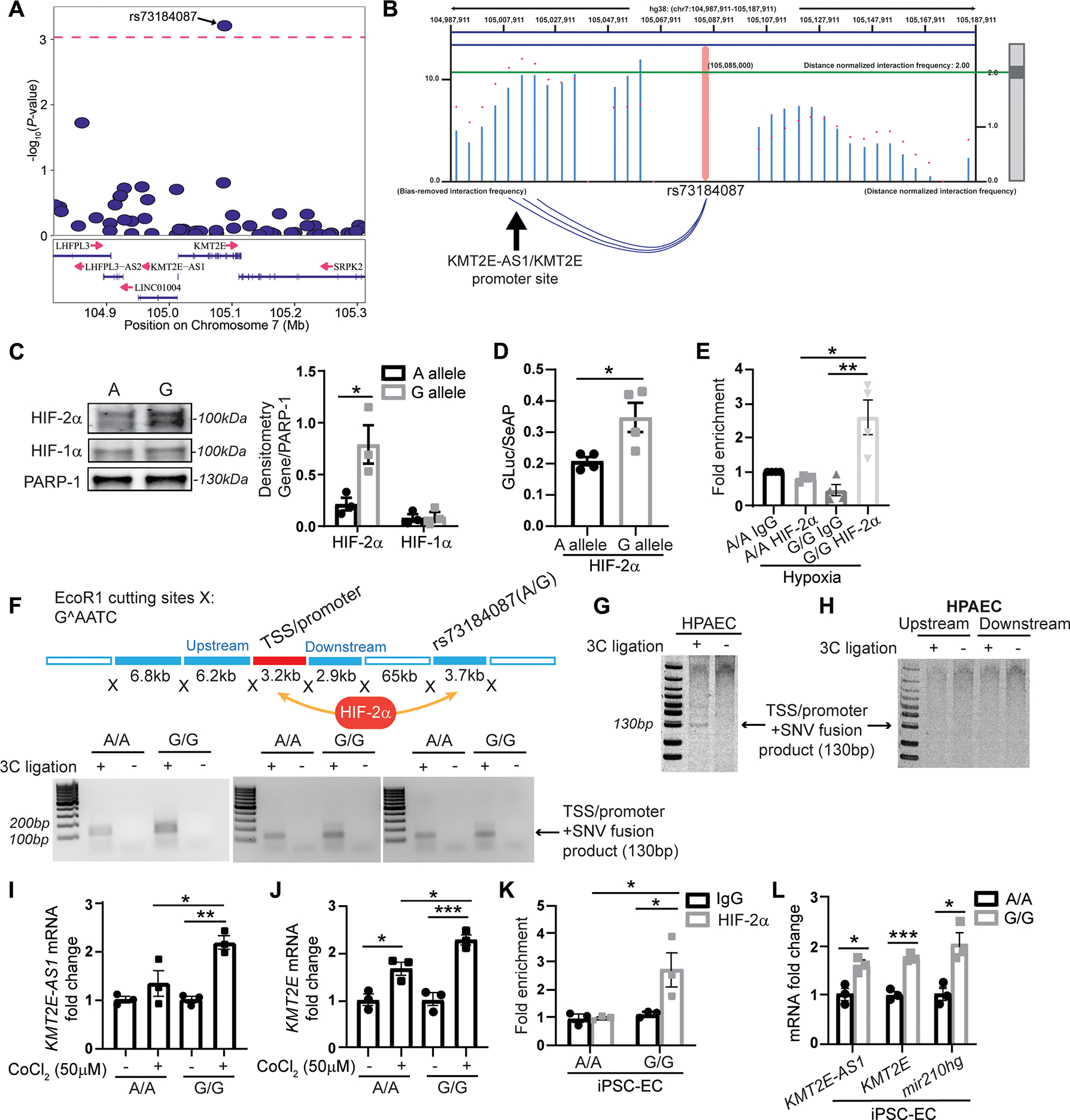
(A) among 883 genotyped and imputed SNVs in the PAH discovery cohort (table S5) within and flanking (+/−200 kb) the lncRNA KMT2e locus, SNVs (table S6) are displayed with predicted HIF-2α binding to either the minor or major SNV allele. independent effective SNV test count was calculated at 53.84 (46). Of those, SNV rs73184087 ranks the highest and meets the P value threshold of 0.00093 (as indicated by the dashed line on the plot). (B) high throughput chromatin conformation capture (HI-C) in lung tissue (29) displays long range interactions between SNV rs73184087 and the transcription start site/promoter region of KMT2E-AS1/KMT2e (as indicated by the blue arcs below the graph). A distance-normalized frequency (magenta dots) greater than the threshold of 2.0 by default (green line) defines a significant interaction with a SNV. (C) SNV binding of HIF-2α and HIF-1α is compared across the SNV a versus G allele (n = 3; *P < 0.05, unpaired Student’s t test; data represent mean ± SEM). (D) Luciferase activity in protein lysates of human embryonic kidney 293T cells transfected with a constitutively active HIF-2α plasmid and a luciferase reporter plasmid carrying the lncRNA-KMT2E promoter and SNV rs73184087 (A versus G allele) (n = 4; *P < 0.05, unpaired Student’s t test; data represent mean ± SEM). Luciferase activity is normalized to constitutively secreted alkaline phosphatase (GLuc/SeAP). (E) ChIP qPCR of HIF-2α binding to SNV in hypoxic transformed lymphocytes from patients with WSPH group 1 PAH carrying SNV rs73184087 (G/G) versus (A/A) genotypes (table S2) (n = 4; *P < 0.05, **P < 0.01, Kruskal-Wallis test followed by dunn’s post hoc analysis; data represent mean ± SEM). (F) In a chromatin conformation capture (3c) assay (top diagram) with three pairs of matched transformed lymphocytes (A/A versus G/G) from patients with WSPH group 1 PAH, PCR is used to detect transcription start site (TSS)/promoter + SNV fusion products indicative of an interaction between SNV rs73184087 and the lncRNA KMT2E promoter. (G and H) detection of a lncRNA-KMT2E promoter and SNV fusion product (G) by 3c assay in human PAECs versus detection of negative control ligation products representing SNV interactions upstream or downstream of the promoter (h). (I and J) KMT2E-AS1 (I) and KMT2E (J) transcripts in transformed lymphocytes carrying A/A or G/G genotype with HIF-α induction by cobalt chloride (50 μM) by RT-qPCR (n = 3; *P < 0.05, **P < 0.01, ***P < 0.001, two-way ANOVA followed by Bonferroni’s post hoc analysis; data represent mean ± SEM). (K) ChIP-qPCR of HIF-2α binding with SNV in inducible pluripotent stem cell differentiated endothelial cells (IPSC-ECS) carrying G/G versus A/A genotype (n = 3; *P < 0.05, two-way ANOVA followed by Bonferroni’s post hoc analysis; data represent mean ± SEM). (L) KMT2E-AS1, KMT2E, and miR210hg transcripts in IPSC-ECS carrying A/A versus G/G genotype by RT-qPCR (n = 3; *P < 0.05, ***P < 0.001, unpaired Student’s t test; data represent mean ± SEM).
