Fig. 5: Iron overload in BED.
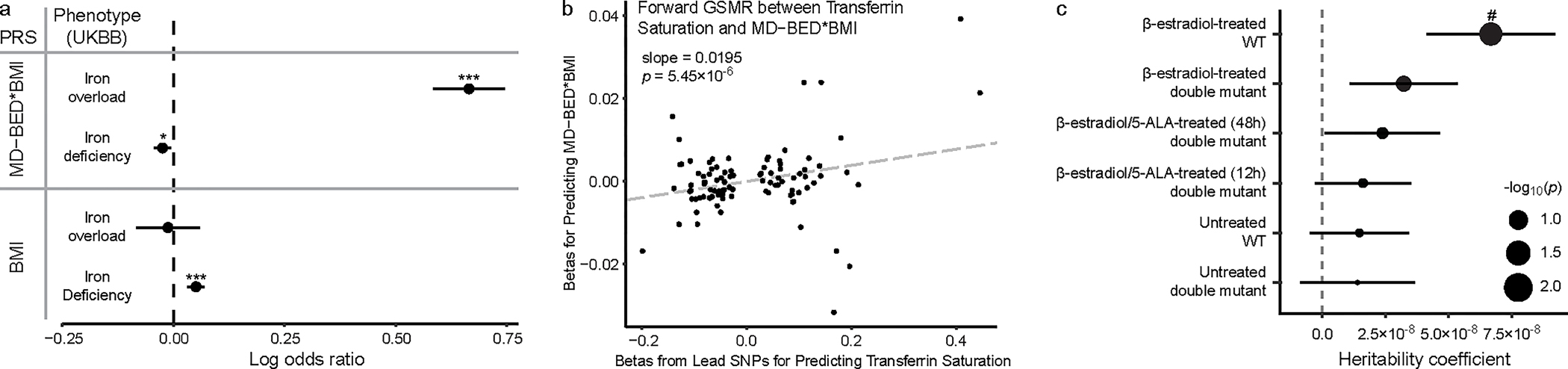
a, PRS associations between EUR-MD-BED*BMI and EUR-BMI GWAS and iron overload (n = 790 cases, n = 385,100 controls) and iron deficiency (n = 11,247 cases, n = 374,643 controls). PRS scores and iron phenotypes are on the y axis. The coefficients, as log odds ratios (mean ± s.e.m.), from the logistic regression for PRS predictors are on the x axis. EUR-MD-BED*BMI PRS predicts iron overload (p = 1.62×10−60) and iron deficiency (p = 0.01). EUR-BMI PRS predicts iron deficiency (p = 1.03×10−7) but not iron overload (p = 0.73). *p < 0.05. ***p < 0.001. b, Scatter plot with generalized linear regression from GSMR between lead SNPs from the transferrin saturation GWAS from deCODE and INTERVAL and EUR-MD-BED*BMI. Transferrin saturation lead SNP betas are on the x axis. EUR-MD-BED*BMI betas are on the y axis. P values from GSMR are from a two-sided Z-test. c, Enrichment of BED risk variant-homologs in open chromatin regions (OCR) in wild type (WT) and heme-deficient mutant murine erythroid cells treated with β-estradiol and/or 5-aminolevulinic acid hydrochloride (5-ALA) (β-estradiol-treated WT: n = 1,010,459 OCR, p = 0.005; β-estradiol-treated double-mutant: n = 1,263,093 OCR, p = 0.07; β-estradiol/5-ALA-treated (48hr) double mutant: n = 1,229,810 OCR, p = 0.15; β-estradiol/5-ALA-treated (12hr) double mutant: n = 1,229,810 OCR, p = 0.20; Untreated WT: n = 1,488,490 OCR, p = 0.23; Untreated double mutant: n = 1,001,591 OCR, p = 0.27). Cell lines are on the y axis. Heritability is on the x axis. Positive coefficients signify enriched heritability. Dot size reflects negative log10 of uncorrected P value (−log10p) from a two-sided LD Score regression Z-test. Error bars indicate standard errors from LD Score regression mean estimates. #p < 0.05 after FDR correction.
