Figure 1. PI3K/AKT pathway inhibitors acting synergistically with an ATRi in PARPi-resistant BRCA2m HGSOC cells.
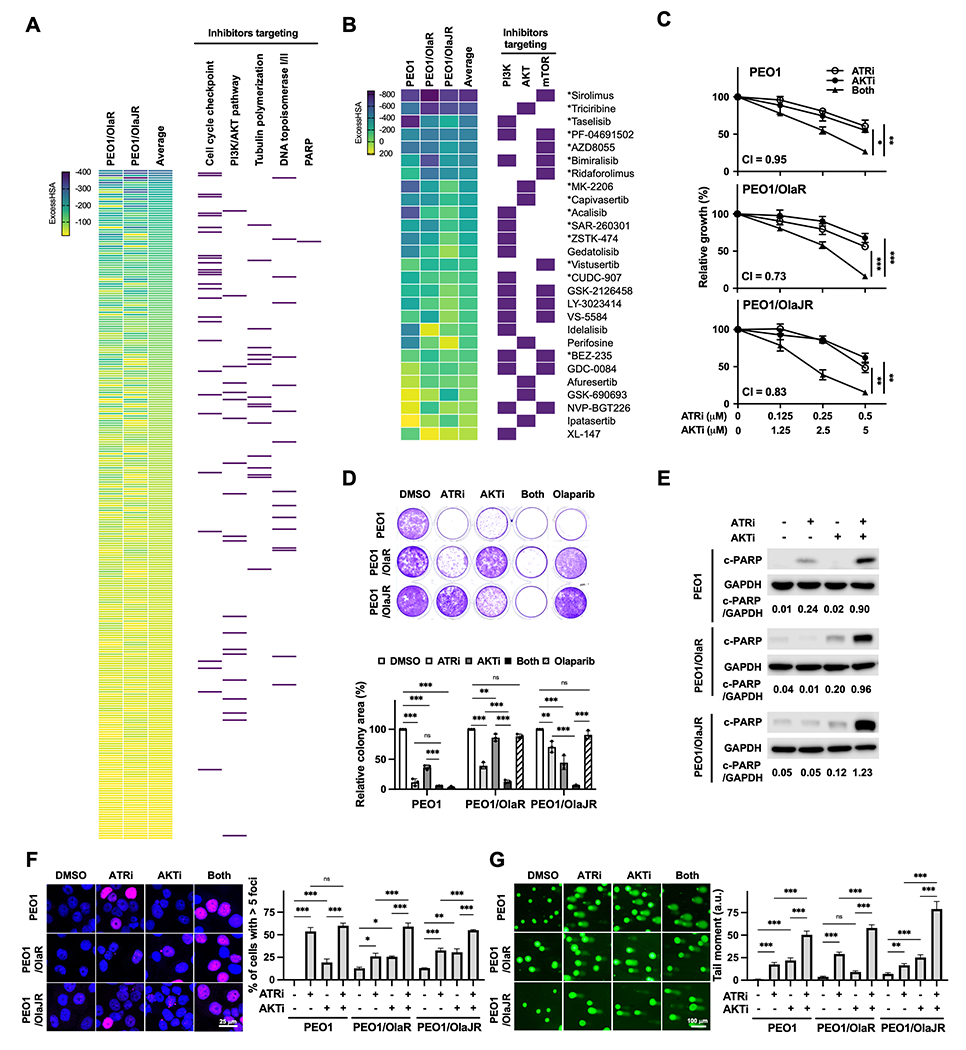
A, Hierarchal view of 6 × 6 initial drug combination screening in PARPi-resistant (PEO1/OlaR and PEO1/OlaJR) BRCA2m HGSOC cells. Drugs that were found to be synergistic with ATRi ceralasertib (ExcessHSA < −20) were ranked using the average ExcessHSA values; more negative ExcessHSA values indicate greater potency. Purple rectangles on the right highlight the agents from key mechanistic classes, including inhibitors targeting cell cycle checkpoint, PI3K/AKT pathway, topoisomerase I/II, tubulin polymerization, and PARP. B, 10 × 10 matrix screening of PI3K/AKT pathway inhibitors and ATRi ceralasertib in PARPi-sensitive (PEO1) and PARPi-resistant (PEO1/OlaR and PEO1/OlaJR) BRCA2m HGSOC cells. Purple rectangles on the right highlight drugs targeting PI3K isoform, AKT, and mTOR. * Indicates the synergy seen in all cell lines. C, Cell growth was assessed using XTT assays (n = 4). Cells were treated with ATRi ceralasertib and/or AKTi capivasertib at indicated doses for 5 days. Combination index (CI) values <1 indicate synergism. D, Long-term survival was evaluated by colony-forming assays (n = 3). Cells were treated with ATRi (0.5 μM) and/or AKTi (5 μM) and grown for 12 days. E-G, Cells were treated with ATRi (1 μM) and AKTi (10 μM) for 48 hours. E, Cell apoptosis effect was assessed by immunoblotting of cleaved PARP (c-PARP). GAPDH was used as a loading control. Densitometric values of c-PARP relative to GAPDH are shown. F-G, DNA damage was examined by immunofluorescence staining for γH2AX foci (n = 3) (F) and alkaline comet assay (n = 3) (G). F, Representative images of γH2AX foci (pink) and nuclei (DAPI, blue) (left). The percentage of cells with > 5 γH2AX foci representing cells with DNA damage is plotted (right). G, Representative images of comet assays are shown (left). The tail moment, including the product of the tail length and the fraction of total DNA, is plotted (right). Data from C, D, F, G were analyzed using one-way ANOVA test and shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
