Figure 6. AKT1 resolves R-loop formation by directly interacting with DHX9 via its kinase domain.
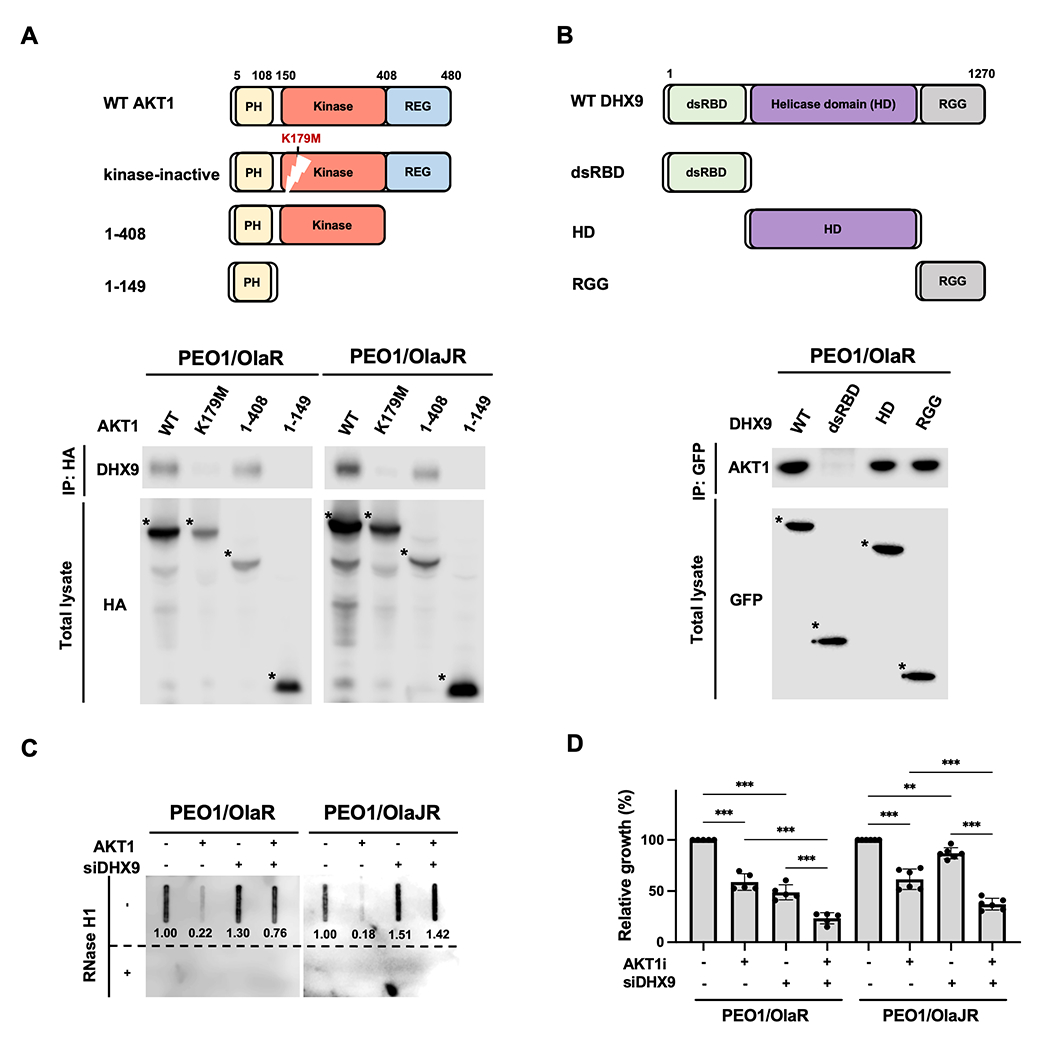
A-B, Schematic representations of AKT1, DHX9 and their serial-deletion mutants (top). Co-IP was performed to study the interaction between DHX9 and AKT1 (bottom). PARPi-resistant HGSOC cells were transfected with (A) HA-tagged full-length or deletion mutants of AKT1 or (B) GFP-tagged full-length or deletion mutants of DHX9. Cell lysates were collected for performing a co-IP assay (n = 3). Bound AKT1 or DHX9 proteins were analyzed by immunoblotting. The full-length or deletion mutants of AKT1 and DHX9 were detected (indicated as *). C, Dot-blot analysis of R-loops. Genomic DNAs were collected from PARPi-resistant cells transfected with siDHX9 and/or GFP-AKT1 (AKT1) for 72 hours. Ratios relative to control group are shown. Bottom panels show the same analysis using genomic DNAs after RNase H1 treatment as control. D, PARPi-resistant HGSOC cells were transfected with siRNAs against DHX9 or scramble controls for 48 hours and then treated with AKTi for another 48 hours. Cell growth was measured by XTT assay (n = 5). Data were analyzed using one-way ANOVA test and shown as mean ± SEM. **, P < 0.01; ***, P < 0.001.
