Figure 1.
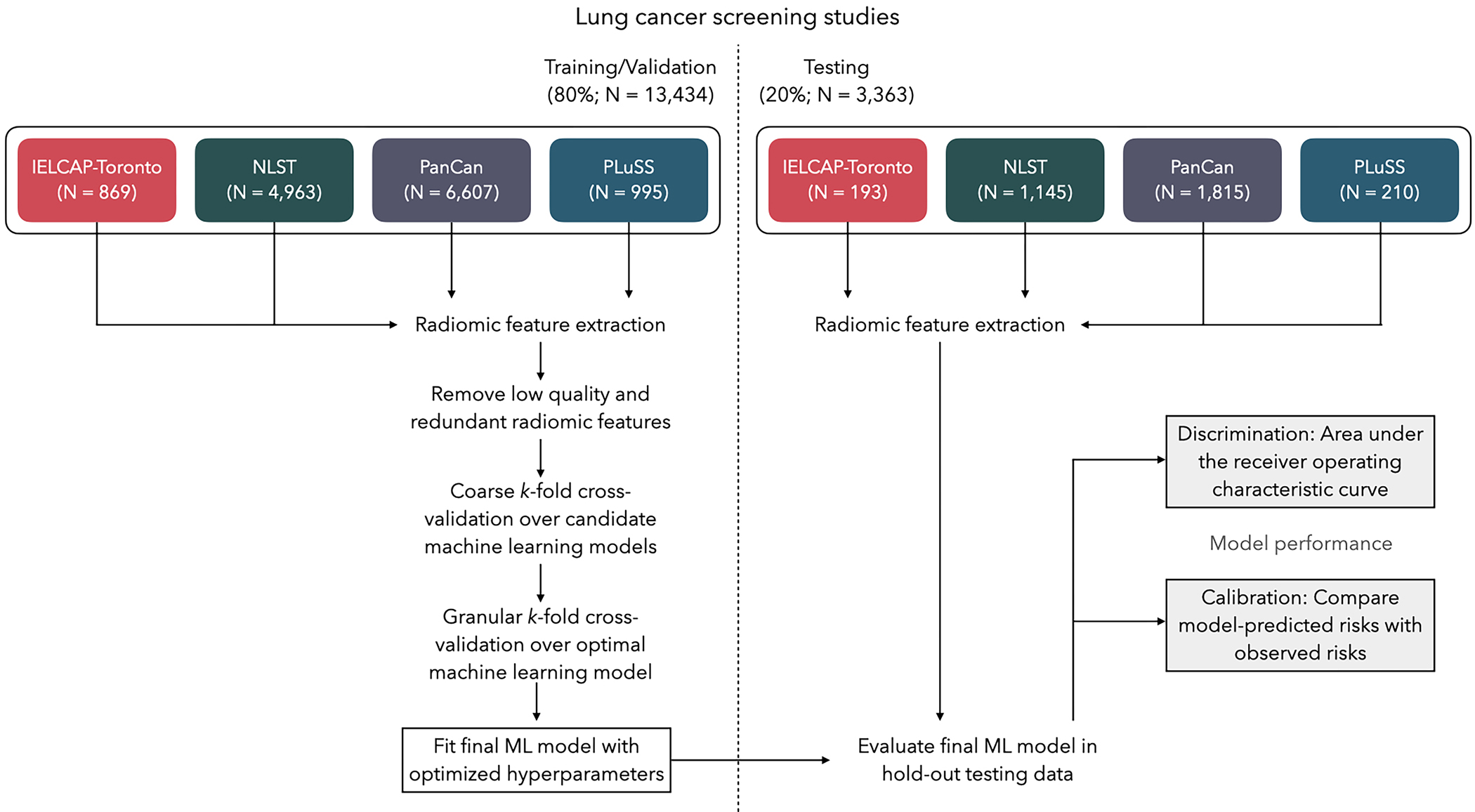
Schematic for the analytic framework used in this study. Data were partitioned into training/validation and testing splits using group-based random sampling to ensure all nodules for a participant were in a single set to avoid data leakage. Radiomic features were extracted and subject to filtering to exclude low-quality and highly-redundant features. K-fold cross-validation was performed to identify the optimal machine learning (ML) model and the optimal set of hyperparameters. The final ML model was fitted to the entire training data set and tested for out-of-sample performance in the hold-out test data; discrimination and calibration performance metrics are reported.
