Figure 2.
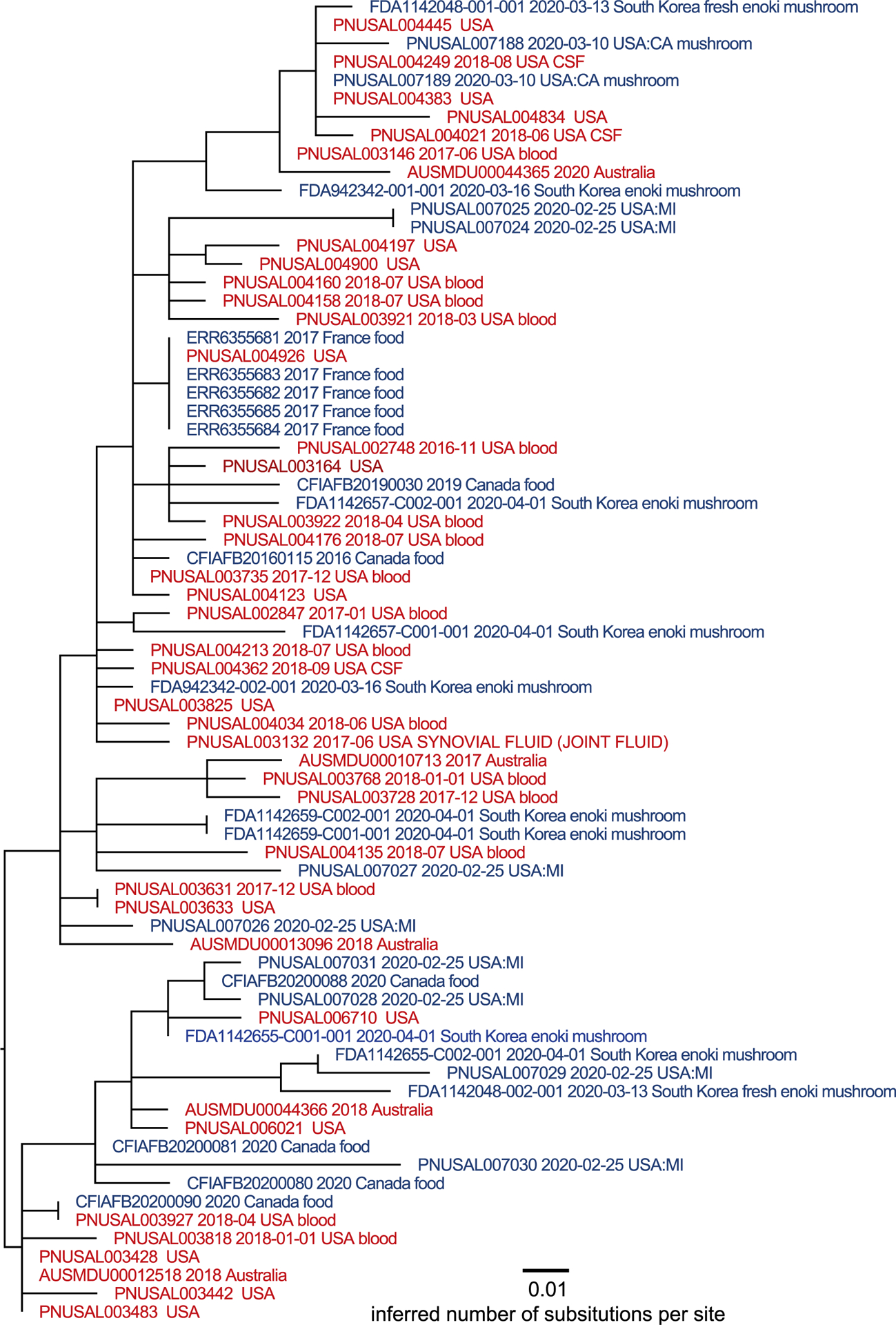
Maximum-likelihood phylogenetic tree inferred via GARLI and a SNP matrix constructed by the CFSAN SNP Pipeline. Tips are color coded as to whether the isolate was collected from a clinical specimen (red) or product sample (blue). Metadata includes isolation date and isolation source in the NCBI Pathogen Database where available for the isolate*. *Single isolate collected by CFIA unrelated to the larger cluster of isolates by WGS. This isolate can be found at https://www.ncbi.nlm.nih.gov/pathogens/tree#Listeria/PDG000000001.2941/PDS000024856.160?key=4qlNyg9jYqcV9ViU95REyULDp6vK-41h91iOKeRJthP_ULcf9H67&labels=epi_type,collection_date,target_creation_date,geo_loc_name,isolation_source,strain,accession.
