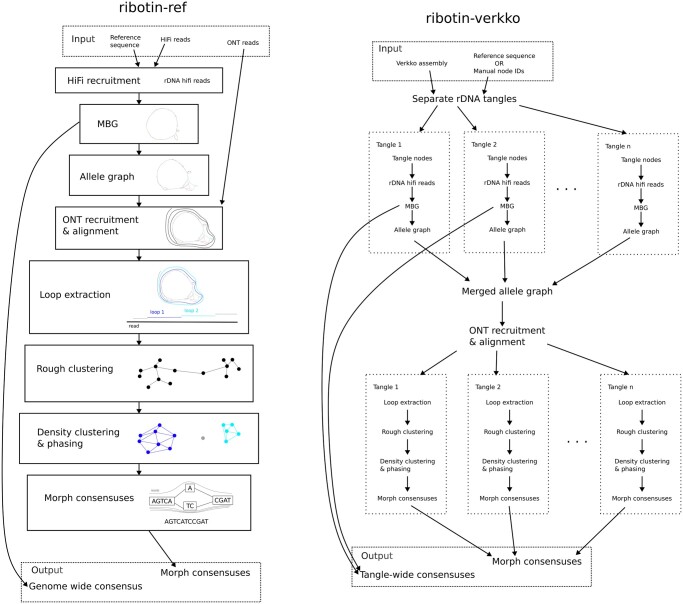
Figure 1.
Overview of ribotin. Left: the reference based ribotin-ref mode. A reference sequence is used to recruit HiFi reads, which are then assembled with MBG and processed into the allele graph which represents all variation present within the rDNA. Then, ONT reads are recruited based on the k-mers of the allele graph and aligned to the allele graph. Loops are extracted from the alignments, which are then clustered to morphs. Finally, a consensus sequence is found for each cluster. Right: the verkko based ribotin-verkko mode. A verkko assembly is required, along with either a reference sequence used for detecting rDNA tangles or a manual assignment of node IDs to rDNA tangles. The HiFi reads are assigned to rDNA tangles based on their locations in the verkko assembly. Then, a pipeline similar to ribotin-ref is used per tangle. The ONT reads are recruited and aligned simultaneously to all allele graphs but otherwise the steps are the same as ribotin-ref.