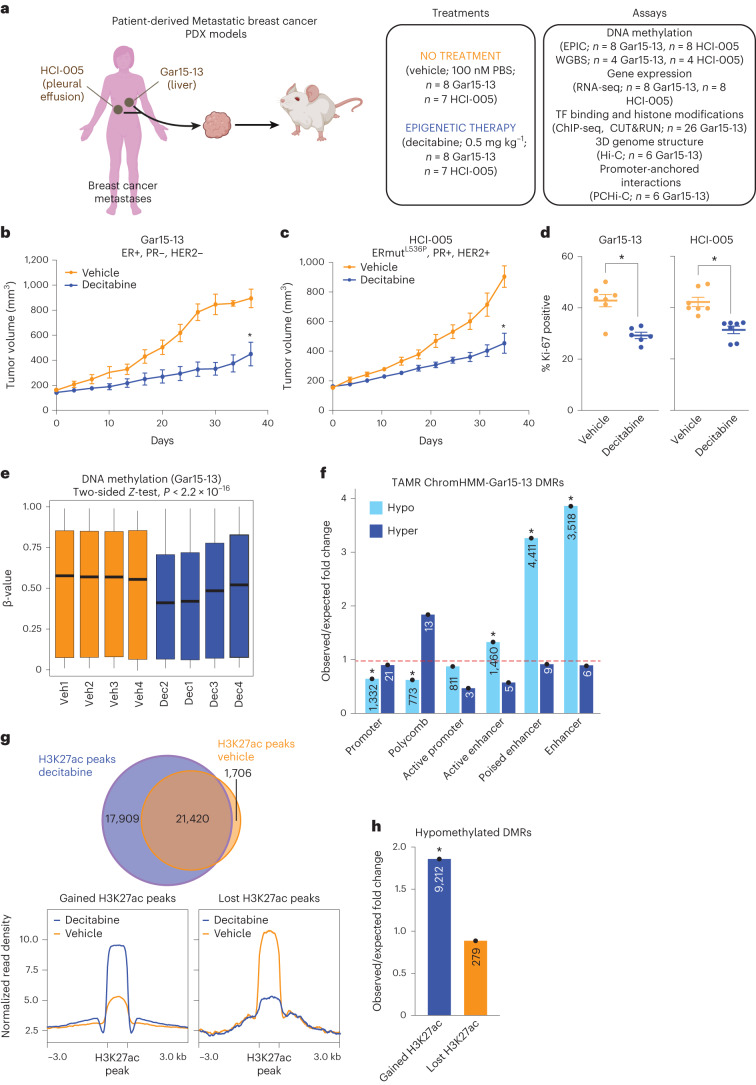
Fig. 1. Decitabine inhibits tumor growth and induces widespread DNA hypomethylation.
a, Schematic of study design. Created with Biorender.com. WGBS: whole-genome bisulfite sequencing; TF, transcription factor b, Gar15-13 PDX growth curves for vehicle-treated (100 nM PBS, n = 7 mice) and decitabine-treated (0.5 mg kg–1, n = 7 mice) tumors. Data are represented as mean ± s.e.m. and analyzed using a two-tailed, unpaired Student’s t-test at the ethical or experimental endpoint. *P < 0.001. Endpoint test details are t = 5.678, df = 8, P = 0.0009. c, HCI-005 PDX growth curves for vehicle-treated (100 nM PBS, n = 8 mice), and decitabine-treated (0.5 mg kg−1, n = 7 mice) tumors. Data are represented as mean ± s.e. and analyzed using a two-tailed, unpaired Student’s t-test at the ethical or experimental endpoint. *P < 0.001. Endpoint test details are t = 5.231, df = 9, P = 0.0001. d, Ki-67 positivity at endpoint in Gar15-13 and HCI-005 PDXs. Data were analyzed using a two-tailed, unpaired Student’s t-test. *P < 0.001. Endpoint test details are t = 4.748, df = 11, P = 0.0006 and t = 4.698, df = 12, P = 0.0005 for Gar15-13 and HCI-005, respectively. e, Distribution of DNA methylation for vehicle-treated and decitabine-treated Gar15-13 PDXs (n = 4 biological replicates each). Box plots show median, interquartile range and maximum–minimum. Data were analyzed using the two-sided Z-test. f, O/E fold change enrichment of DMRs in Gar15-13 decitabine compared to vehicle across TAMR ChromHMM regulatory regions. *P < 0.001 (permutation test). Numbers located within each specific region are presented in the respective column. g, Overlap of consensus H3K27ac peaks between vehicle-treated and decitabine-treated Gar15-13 PDXs (n = 3 biological replicates each). Average signal intensity of H3K27ac at gained and lost H3K27ac binding sites in Gar15-13 PDXs. h, O/E fold change enrichment of hypomethylated DMRs in Gar15-13 decitabine compared to vehicle across gained and lost H3K27ac peaks. *P < 0.001 (permutation test). The numbers located within each specific region are presented in the respective column.