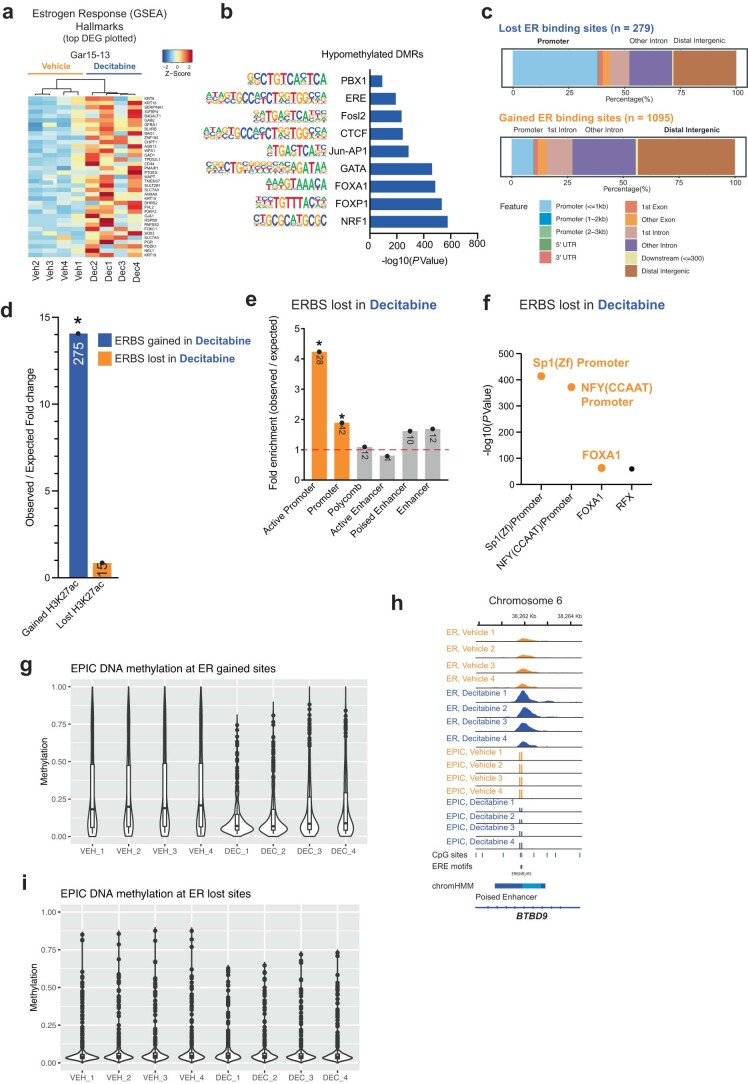
Extended Data Fig. 6. Alterations to Estrogen Receptor binding upon Decitabine treatment.
(a) RNA-seq heatmap of Decitabine-induced changes in expression of genes belonging to the Estrogen Response (GSEA) Hallmarks. Top differentially expressed genes plotted (FDR < 0.05). (b) Transcriptions factor motifs enriched at hypomethylated DMRs between Vehicle- and Decitabine-treated Gar15-13 PDXs compared to matched random regions across the genome. Only motifs with binomial P value < 0.05 are shown. (c) RefSeq annotation of Vehicle vs. Decitabine lost and gained ERBS in Gar15-13. (d) Observed over expected fold change enrichment of gained ER binding sites (ERBS) and gained and lost H3K27ac binding sites. * P value < 0.001 (permutation test). Size of the overlap is presented in the respective column. (e) ChromHMM (TAMR) annotation (*P value < 0.001, permutation test) of ER binding sites lost with Decitabine treatment compared to matched random regions across the genome. Size of the overlap is presented in the respective column. (f) Motifs enriched at ERBS lost with Decitabine treatment compared to matched random regions generated from ERE binding motifs across the genome. Only motifs with binomial P value < 0.05 are shown. (g) DNA methylation levels at gained ERBS in Decitabine-treated (n = 4 biological replicates) and Vehicle (n = 4 biological replicates) PDX Gar15-13 PDXs. Box plots show median, inter-quartile range and maximum/minimum log fold change. (h) Browser snapshot of ER ChIP-seq and EPIC DNA methylation (Vehicle and Decitabine-treated PDXs, n = 4 biological replicates each), showing concomitant gain of ER binding and loss of DNA methylation at enhancer of ER target gene BTBD9. (i) DNA methylation levels at lost ERBS in Decitabine-treated (n = 4 biological replicates) and Vehicle (n = 4 biological replicates) Gar15-13 PDXs. Box plots show median, inter-quartile range and maximum/minimum log fold change. DMR, differentially methylated region. ERBS, ER binding site.