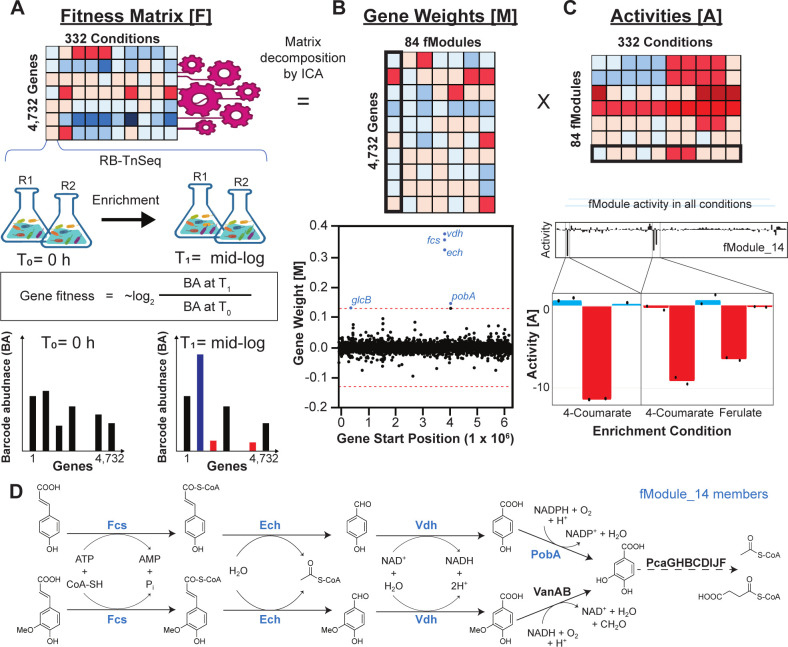
Fig 1.
Decomposition of RB-TnSeq multivariate gene fitness profiles using ICA. (A) Schematic of the general RB-TnSeq approach. A randomly barcoded transposon insertion library in P. putida is cultivated in a baseline (“time zero”, T0) condition and then enriched in a selective condition to mid-log (T1). Normalized barcode counts, which are indicative of specific mutant frequency in the population, are enumerated before and after enrichment to calculate a fitness value for each gene disruption. Genes with larger fitness magnitudes are expected to play a more important functional role for a given condition. In ICA, (A) fitness values, , from RB-TnSeq experiments were decomposed into a matrix of (B) gene weights, , and a matrix of (C) condition-specific activities, . In , genes with outlier weight values were grouped together into a single functional group, termed a “functional module”, which we abbreviate as “fModule”. In , the activity profile of an fModule characterizes the related biological functions of its member genes in each RB-TnSeq enrichment condition. Data from fModule_14 is provided as an example of ICA fModule gene weight and fModule activity outputs. (D) fModule_14 contains several genes involved in hydroxycinnamate metabolism. The negative activity of fModule_14 for conditions with 4-coumarate or ferulate as a sole carbon source in panel C is consistent with the essential role of its member gene products in catabolism of these substrates.