Extended Data Fig. 7 |. Geneformer boosted predictions in a diverse panel of downstream tasks.
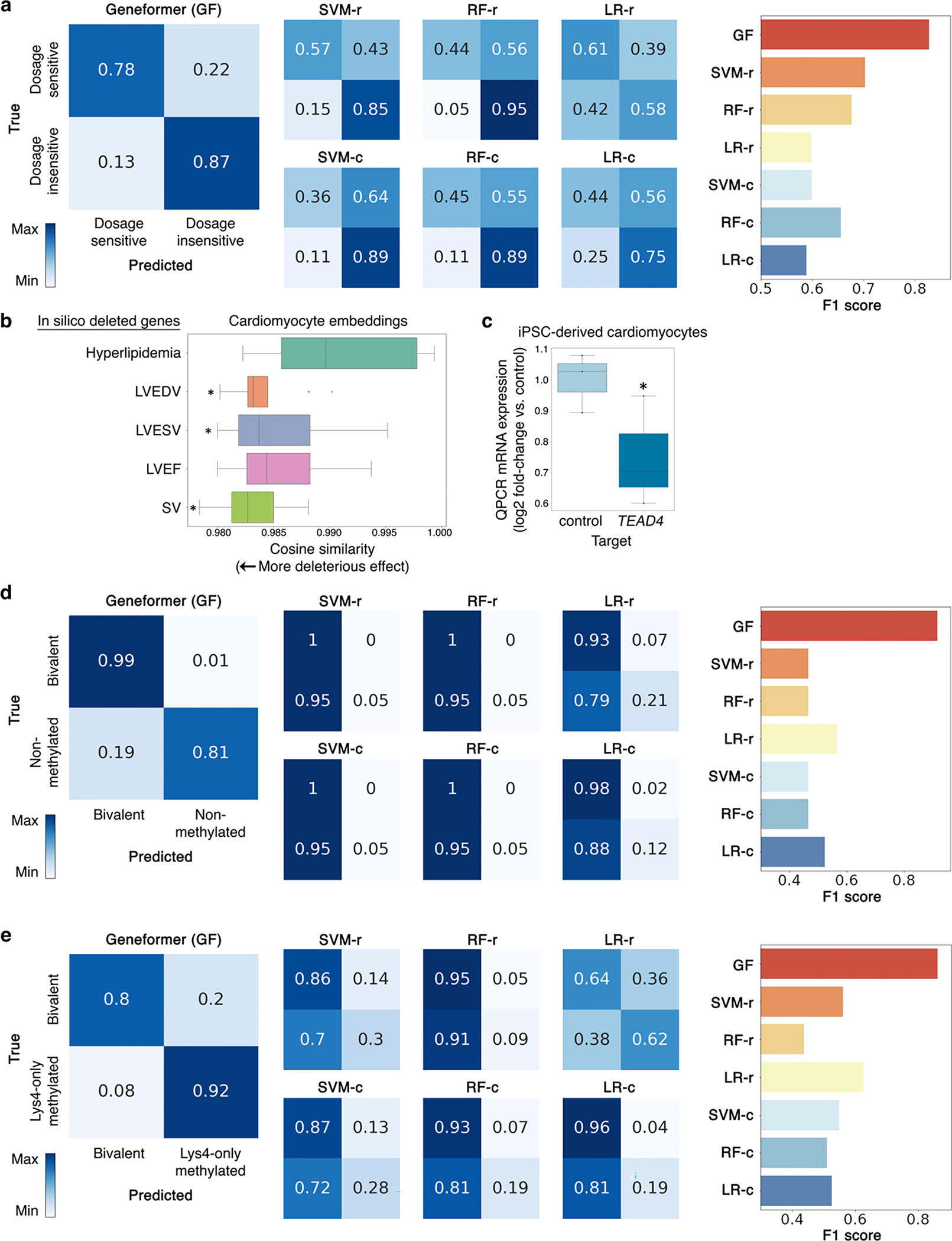
a, Confusion matrices and F1 score for Geneformer predictions vs. alternative methods (as described in Fig. 2a) for downstream task of distinguishing dosage-sensitive vs. insensitive transcription factors. b, Effect on cardiomyocyte embeddings from in silico deletion of genes linked by prior transcriptome-wide association study (TWAS)-prioritized GWAS24 to cardiac MRI traits relevant to cardiac pathology (left ventricular (LV) end diastolic volume (EDV), LV end systolic volume (LVESV), LV ejection fraction (LVEF), and stroke volume (SV)) compared to in silico deletion of control cardiac disease genes expressed in cardiomyocytes but whose pathology occurs in non-cardiomyocyte cell types (hyperlipidemia). (*p<0.05 by Wilcoxon, FDR-corrected; center line=median, box limits=upper and lower quartiles, whiskers=1.5x interquartile range, points=outliers). c, Quantitative PCR (QPCR) data of CRISPR-mediated knockout of TEAD4 in iPSC-derived cardiomyocytes (n=3, *p<0.05 by t-test; center line=median, box limits=upper and lower quartiles, whiskers=1.5x interquartile range, points=experimental replicates). d, Confusion matrices and F1 score for Geneformer predictions vs. alternative methods for downstream task of distinguishing bivalent vs. non-methylated genes (56 highly conserved loci28). e, Confusion matrices and F1 score for Geneformer predictions vs. alternative methods for downstream task of distinguishing bivalent vs. Lys4-only methylated genes (56 highly conserved loci28).
