Extended Data Fig. 10 |. Geneformer fine-tuned cardiomyocyte embeddings clustered by phenotype.
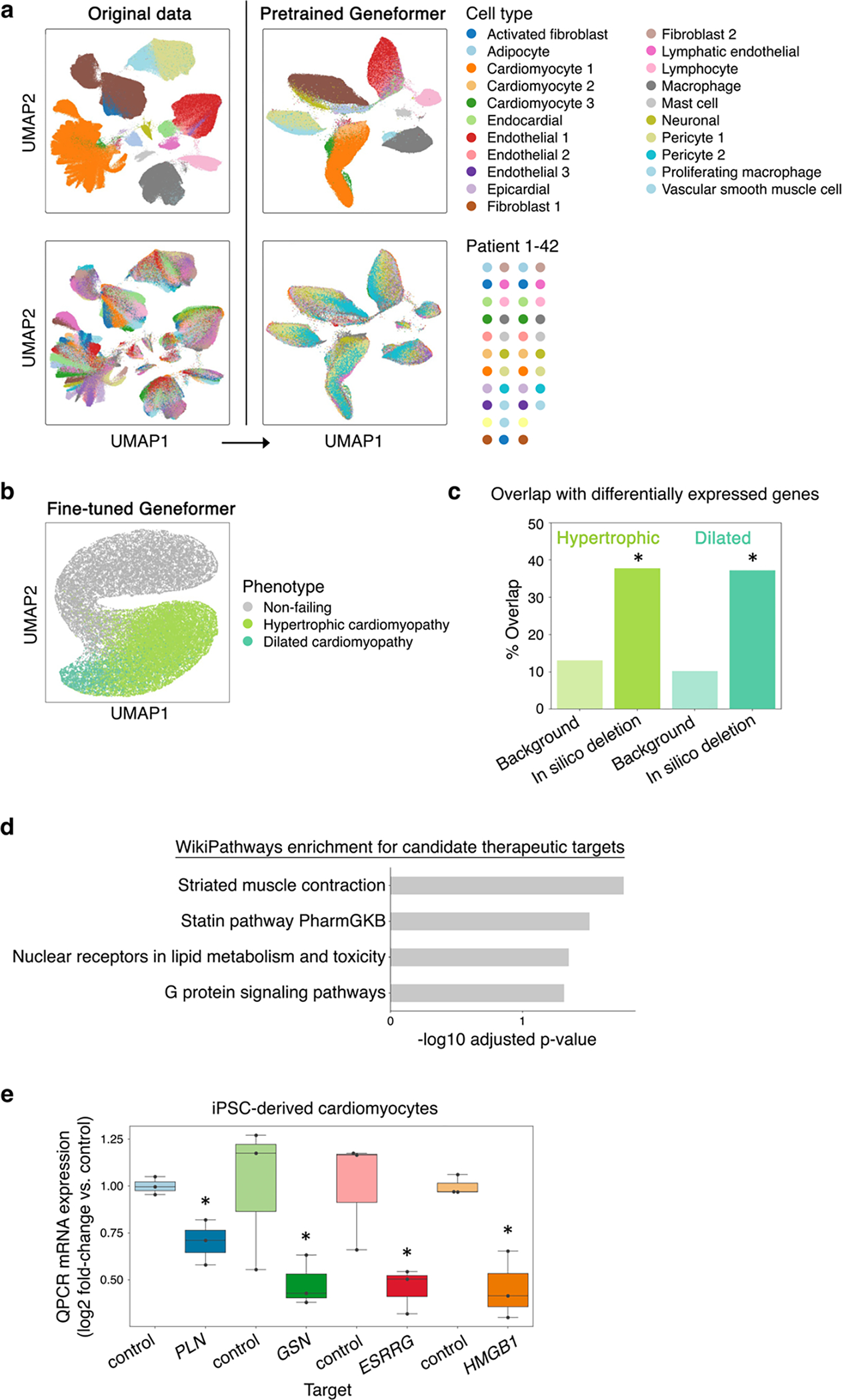
a, While original data (left) was highly affected by patient batch effect, cell embeddings generated by pretrained Geneformer (right) (without fine-tuning) clustered primarily by cell type. b, UMAP of cardiomyocyte embeddings from the model fine-tuned to distinguish cardiomyocytes in non-failing hearts from cardiomyocytes in patients with hypertrophic or dilated cardiomyopathy. c, Gene sets significantly associated with hypertrophic or dilated cardiomyopathy states by Geneformer in silico deletion disease modeling significantly overlapped with genes differentially expressed in those respective disease states (differentially expressed vs. non-failing) compared to the overlap of those differentially expressed genes with background genes (the remainder of the genes detected in cardiomyocytes that were not significantly associated with hypertrophic or dilated cardiomyopathy by Geneformer disease modeling) (*p<0.05 by X2 test, FDR-corrected). d, Pathway enrichment for genes whose in silico deletion in cardiomyocytes from hypertrophic cardiomyopathy patients significantly shifted embeddings towards the non-failing state and away from the dilated cardiomyopathy state, suggesting candidate therapeutic targets. e, QPCR data of CRISPR-mediated knockout of indicated genes in TTN+/− iPSC-derived cardiomyocytes (n=3, *p<0.05 by t-test). Center line=median, box limits=upper and lower quartiles, whiskers=1.5x interquartile range, points=experimental replicates.
