Extended Data Fig. 1 |. Geneformer transfer learning strategy.
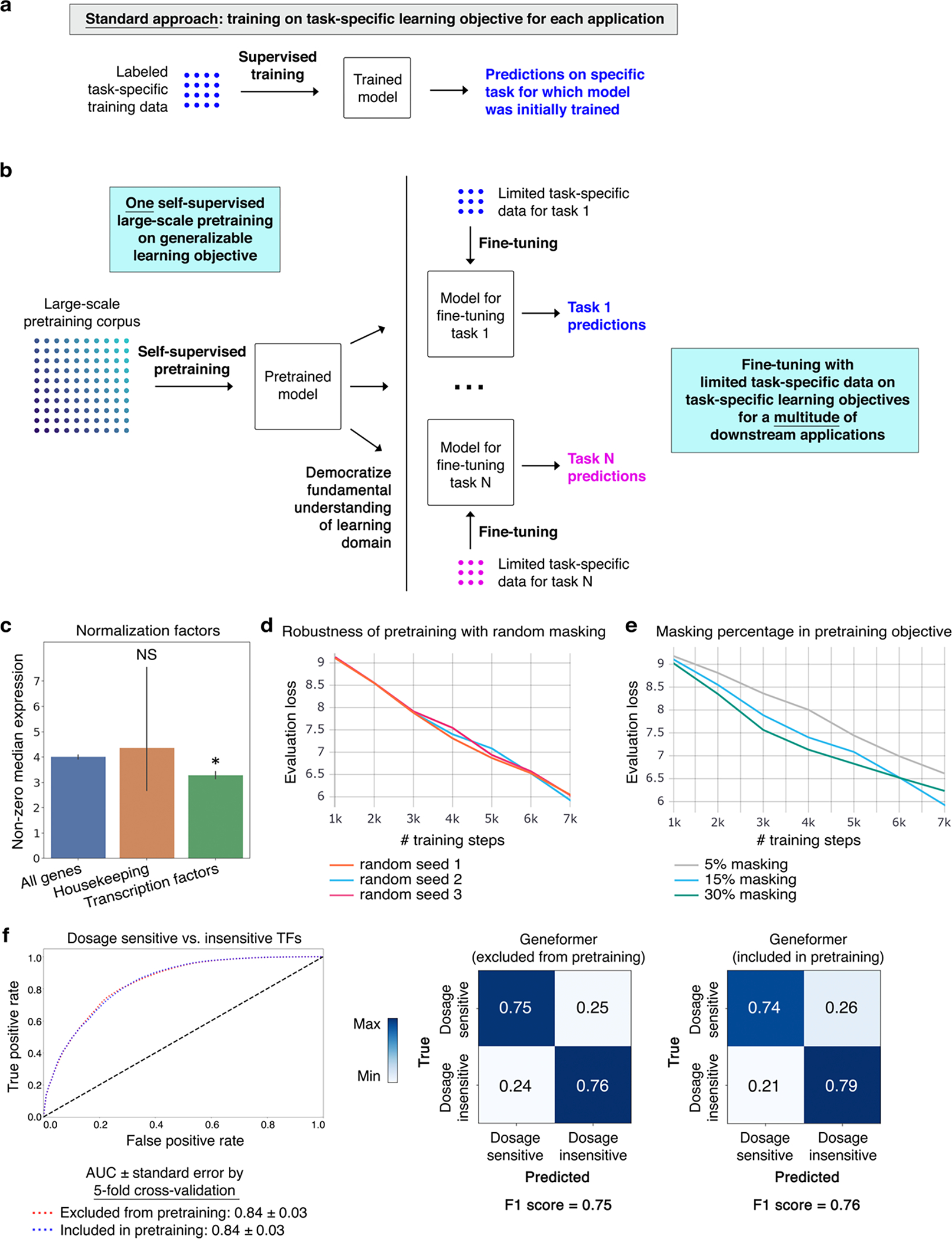
a, Schematic of standard modelling approach, which necessitates retraining a new model from scratch for each new task. b, Schematic of transfer learning strategy. Through a single initial self-supervised large-scale pretraining on a generalizable learning objective, the model gains fundamental knowledge of the learning domain that is then democratized to a multitude of downstream applications distinct from the pretraining learning objective, transferring knowledge to new tasks. c, Transcription factors are normalized by a statistically significantly lower factor (resulting in higher prioritization in the rank value encoding) compared to all genes. Housekeeping genes on average show a trend of a higher normalization factor (resulting in deprioritization in the rank value encoding) compared to all genes (*p<0.05 by Wilcoxon, FDR-corrected; all genes n=17,903, housekeeping genes n=11, transcription factors n=1,384; error bars=standard deviation). d, Pretraining was performed with a randomly subsampled corpus of 100,000 cells, holding out 10,000 cells for evaluation, with 3 different random seeds. Evaluation loss was essentially equivalent in the 3 trials, indicating robustness to the set of genes randomly masked for each cell during the pretraining. e, Pretraining was performed with a randomly subsampled corpus of 100,000 cells, holding out 10,000 cells for evaluation, with 3 different masking percentages. 15% masking had marginally lower evaluation loss compared to 5% or 30% masking. f, Pretraining was performed with a randomly subsampled corpus of 90,000 cells and the model was then fine-tuned to distinguish dosage-sensitive vs. -insensitive transcription factors using 10,000 cells that were either included in or excluded from the 90,000 cell pretraining corpus. Predictive potential on the downstream fine-tuning task was measured by 5-fold cross-validation with these 10,000 cells, demonstrating essentially equivalent results by AUC, confusion matrices, and F1 score. Because the fine-tuning applications are trained on classification objectives that are completely separate from the masked learning objective, whether or not task-specific data was included in the pretraining corpus is not relevant to the downstream classification predictions.
