Extended Data Fig. 2 |. Geneformer was context-aware and robust to batch-dependent technical artifacts.
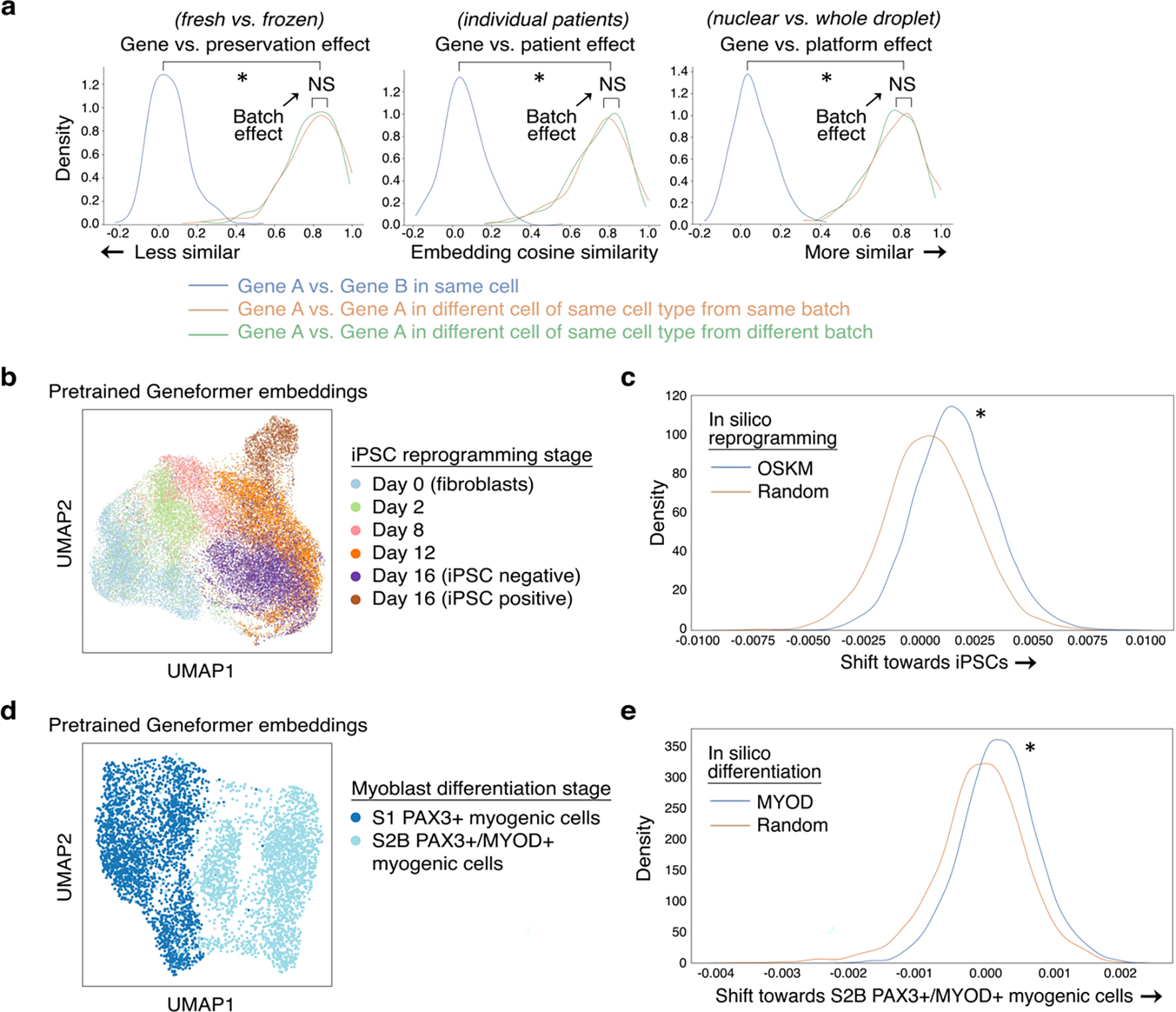
a, Effect of gene versus the indicated batch-dependent technical artifact on pretrained Geneformer gene embeddings (*p<0.05 by Wilcoxon, FDR-corrected; NS: non-significant). We found that the gene embeddings were robust to sequencing platformM11, preservation method13,12, and individual patient variability14. b, UMAP of pretrained Geneformer cell embeddings of cells undergoing iPSC reprogramming appropriately captured temporal trajectory of reprogramming (cell types as annotated by original study15; iPSC negative or positive refers to expression of marker TRA-1–60). Cell embeddings suggested that cells which do not progress to the iPSC state bifurcate into an alternative fate compared to cells that progress to the iPSC state after the day 12 stage. c, Compared to in silico reprogramming with random genes, in silico reprogramming of fibroblasts by artificially adding OCT4, SOX2, KLF4, and MYC (OSKM) to the front of their rank value encodings significantly shifted the gene embeddings from their initial fibroblast state to the embedding of that gene in the iPSC state (*p<0.05 by Wilcoxon). d, UMAP of pretrained Geneformer cell embeddings of cells undergoing iPSC to myoblast differentiation at the earlier S1 (PAX3+) and later S2B (PAX3+/MYOD+) stages (cell types as annotated by original study16). e, Compared to in silico differentiation with random genes, in silico differentiation of the early-stage myogenic cells by artificially adding MYOD to the front of their rank value encodings significantly shifted the gene embeddings from their earlier state to the embedding of that gene in the later MYOD+ myogenic state (*p<0.05 by Wilcoxon).
