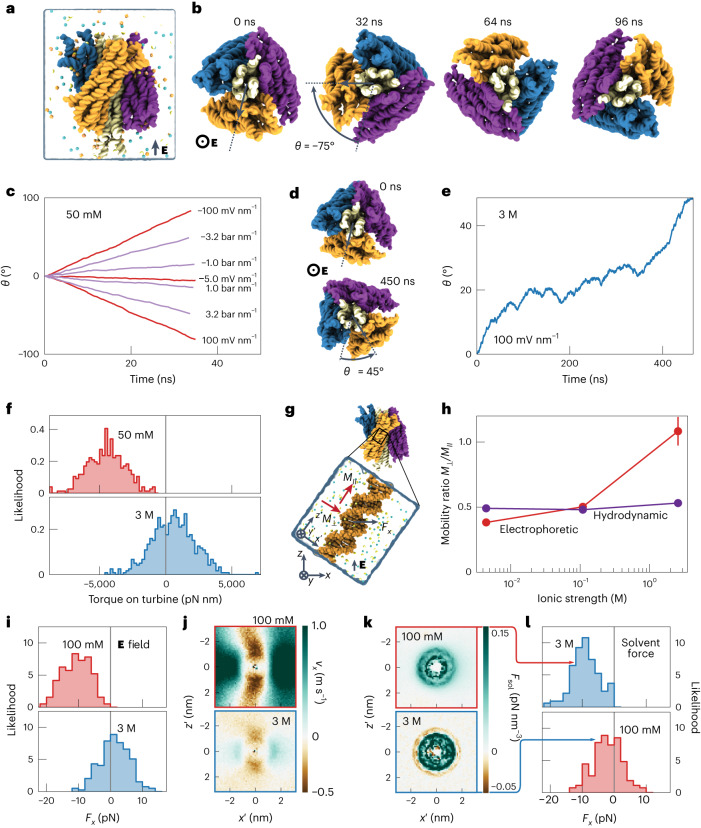
Fig. 4. All-atom MD simulation of a DNA turbine rotation.
a, 4,322,088-atom model of a DNA origami turbine that is depicted using a molecular surface representation (white shaft, multicoloured blades), solvent shown as a semi-transparent surface, and ions (50 mM NaCl, 10 mM Mg2+) shown explicitly. b, Rotation of the turbine driven by electric field (50 mM NaCl). A 100 mV nm−1 field was applied out of the page while restraints prevented the drift and tilting of the turbine. c, Rotation angle of the turbine due to an applied field or pressure gradient. d,e, The same as b,c, but for simulations at 3 M NaCl. A reversal of the rotation direction is observed. f, Torque on the turbine measured by restraining its spin angle. g, Single DNA helix as a minimal model for a turbine blade. A field or pressure gradient is applied along the +z direction. h, Ratio of the electrophoretic and hydrodynamic mobilities for motion perpendicular and parallel to the DNA helical axis observed in simulations of a single DNA helix. Ionic strength is calculated from the average molality observed at distances beyond 6 nm from the helical axis. i, Force on DNA orthogonal to the electric field measured from the single-helix simulations. j, In-plane solvent flow along the x axis in the simulations under an applied electric field. Heat maps depict the average along the helical axis of the DNA. The coordinate system is defined in g. k, Solvent forces extracted from MD simulation under an applied electric field. l, Force on DNA orthogonal to the applied force axis when, as a control, the solvent forces extracted from the low- and high-salt systems are applied to high- and low-salt systems, respectively. The forces were seen to reverse when compared with those in i, proving the causal role of the ion distribution.