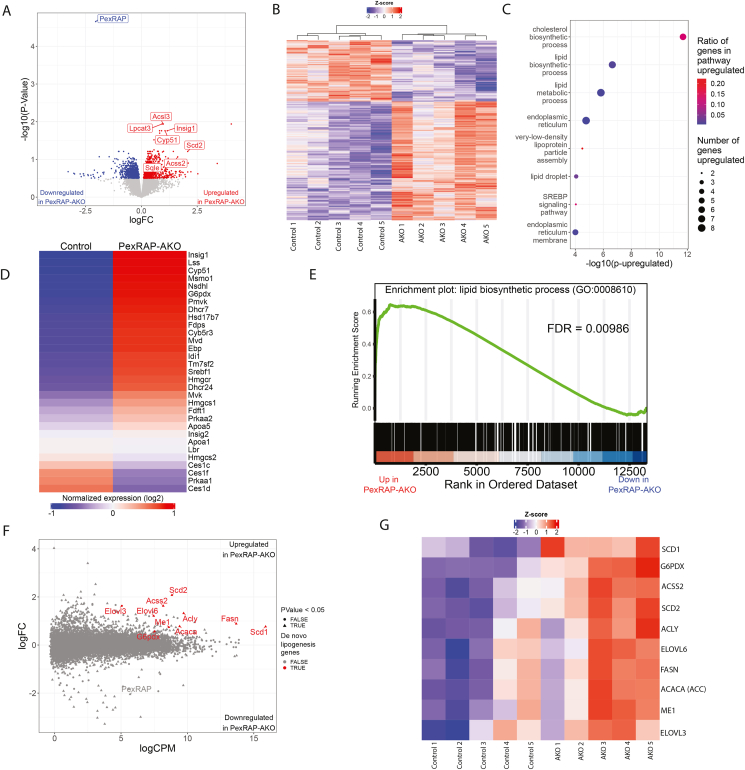
Figure 3.
PexRAP-AKO increases transcription of genes involved in de novo lipogenesis and cholesterol synthesis in iWAT.A) Volcano plot of differentially expressed genes from bulk RNA-sequencing analysis of iWAT from PexRAP and control mice. Several genes that were very significantly increased are highlighted. B) Heatmap of logCPM value-derived Z-scores. C) Gene ontology (GO) pathway enrichment analysis of genes differentially expressed (FDR ≤ 0.05) in PexRAP-AKO mice. D) Heatmap of all genes in the cholesterol biosynthetic process GO term (GO:0006695). E) Enrichment plot of GO:0008610 (lipid biosynthetic process), a top result of an unbiased GSEA analysis. F) MA plot (logFC vs. logCPM) of results of differential expression analysis of RNA-seq data from control and PexRAP-AKO mice (n = 5/group for all). G) Heatmap of key lipid synthetic genes.