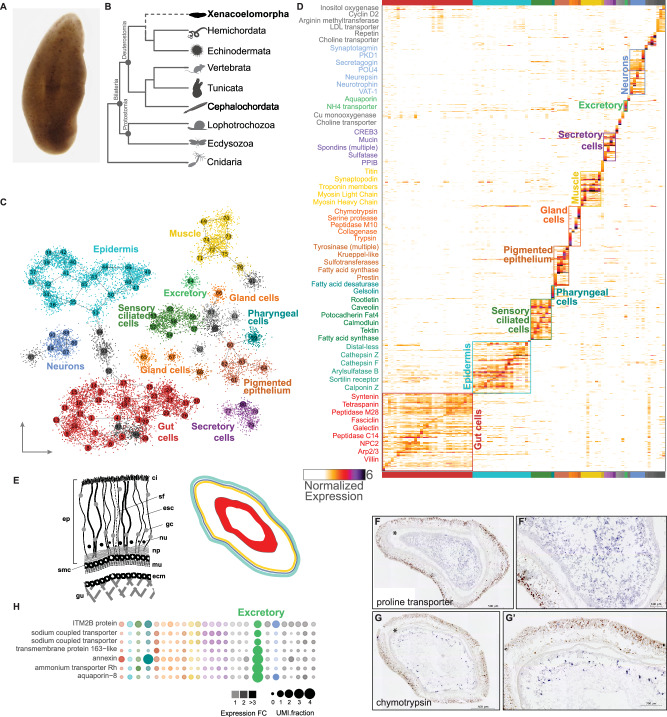
Fig. 1. Xenoturbella single-cell RNA-seq identifies spatially distinct cell types and a minimally complex neural cell type complement.
A A light micrograph of a live Xenoturbella bocki. B Phylogenetic tree showing the recently proposed position of X. bocki as sister to Ambulacraria. C 2D projection of 97 metacells (indicated with numbers) and 12,350 X. bocki cells. D A cell by gene heatmap of cells from C showing expression similarity within the distinct tissue types. E A schematized diagram of a cross-section of Xenoturbella tissues: ci (cilia), sf (supporting filament), esc (epidermal supporting cell), gc (gland cell), nu (nucleus), np (nerve plexus), m (muscle), ecm (extracellular matrix), gu (gut), smc (sub-epidermal membrane complex), ep (epidermis). Right, schema of a horizontal cross-section of a Xenoturbella individual in which endodermal (red), ectodermal (aqua), and mesodermal (yellow) derivatives are indicated. F, G In situ hybridization of cell-type markers identified from the scRNA-seq clustering. F, G are high magnification images of F', G', respectively. H Markers associated with excretion co-localize in an unknown cell type (green). Images used to generate 1b were obtained from phylopic.org.