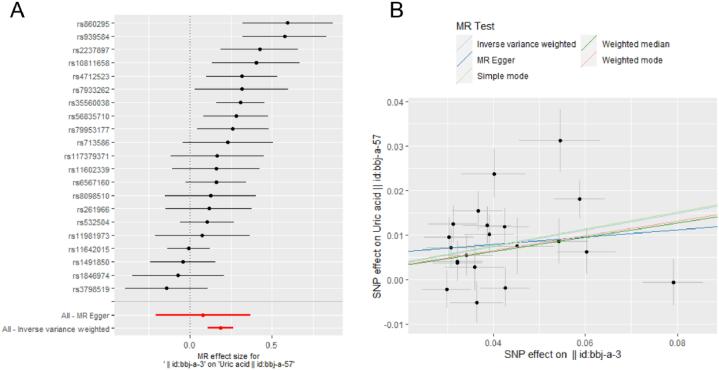
Fig. 2.
Mendelian randomization (MR) analysis from body mass index (BMI) to uric acid (UA). Forest plot (A). Scatter plot (B). In the Forest plot, the black dots represent the logarithm odds ratio (lgOR) of UA increased by the standard deviation (SD) in BMI, which is generated using each single nucleotide polymorphism (SNP) as a separate tool variable. The red dots show the effect sizes of all SNP combinations by different MR methods. Horizontal bars represents 95% confidence interval (CI). In the scatter plot, X axis (SD unit) represents the effect of SNP on BMI, Y axis (lgOR) represents the effect of SNP on uric acid. Each black dot represents a separate SNP, The vertical and the horizontal bars represent 95% CI of the effect size of uric acid and BMI, respectively. Three slopes represent the causal estimation for three MR methods. The light blue slope is the IVW method, the dark blue slope is the MR Egger method, and the green slope is the weighted median method.