Fig. 5.
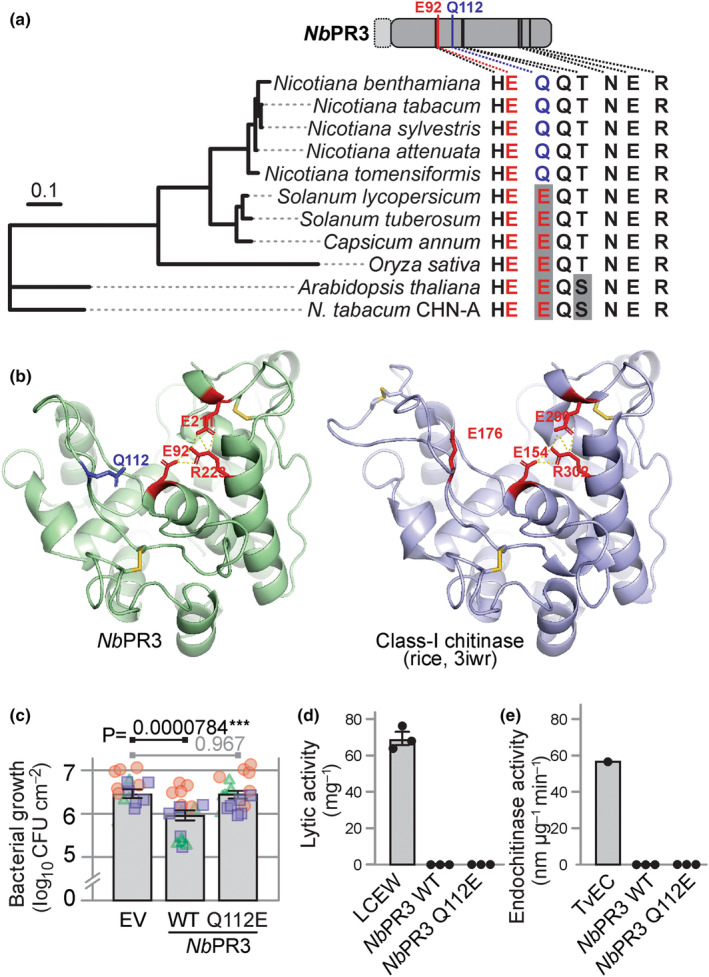
Nicotiana‐specific active site Q112 in NbPR3 is essential for antibacterial activity. (a) Conservation of chitinase‐relevant residues in NbPR3 homologs. The second catalytic glutamate of chitinases (red) is replaced by glutamine (blue) in all NbPR3 orthologs of Nicotiana. NbPR3 orthologs were identified by Blast searches, aligned by ClustalO (Supporting Information Fig. S6), and used to construct a maximum likelihood phylogenetic tree. Chitinase A (CHN‐A) from Nicotiana tabacum was included as the closest related enzyme for which chitinase activity has been demonstrated. The residues relevant for chitinase activity are summarised on the right, showing that the Gln is conserved in Nicotiana and is specific for this genus. (b) Model of NbPR3 structure, compared with rice class‐I chitinase. A model of NbPR3 was generated with SWISS Model, using protein data bank 3iwr (Kezuka et al., 2010) as a template. Catalytically important residues (red), and the noncanonical Q112 (blue), and three cysteine bridges (yellow) are indicated. (c) The Nicotiana‐specific Q112 is essential for antibacterial activity. NbPR3, its Q112E substitution mutant, and the empty vector (EV) control were transiently expressed in Nicotiana benthamiana by agroinfiltration, and 2 d later, the same leaf was infiltrated with 106 CFU ml−1 PtoDC3000(ΔhQ). Bacterial population sizes 3 d later are shown in log10CFU cm−2. Bars show the mean value of 18 biological replicates performed over three separate experiments, and error intervals represent the SE. P‐values were calculated by two‐way ANOVA followed by post hoc comparison using the Dunnett test to examine the effect of NbPR3 expression on bacterial growth. ***, P < 0.001. (d) The Q112E substitution in NbPR3 does not increase lytic activity. Apoplastic fluid (AF) from plants transiently expressing NbPR3 or its Q112E mutant was incubated with Micrococcus lysodeikticus cells. The change in A450 was measured and converted to units μg−1 NbPR3 protein estimated by Coomassie staining. The lysozyme of chicken egg white (LCEW) was included as a positive control. (e) The Q112E substitution in NbPR3 does not increase endochitinase. AF from plants transiently expressing NbPR3 or its Q112E mutant was incubated with 4‐MU‐GlcNac3, and the rate of hydrolysis was measured at 355ex/450em and calculated per μg NbPR3 protein estimated by Coomassie staining. The endochitinase of Trichoderma viride (TvEC) was included as a positive control.
