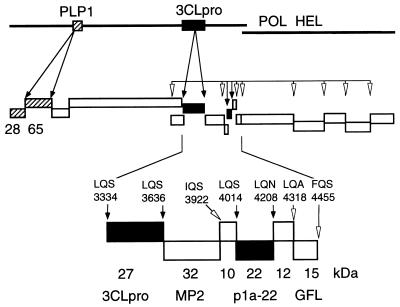
FIG. 6.
MHV gene 1 polyprotein processing. This model is based on known and predicted cleavage sites and proteins identified in cells and in vitro. PLP-1 and proteins known to be cleaved in trans by PLP-1 (p28 and p65) are shown as hatched boxes. 3CLpro and proteins known to be cleaved by 3CLpro (3CLpro and p1a-22) are shown as filled boxes. Other predicted proteins are shown as open boxes. Experimentally confirmed cleavage sites are noted by filled arrows, while putative cleavage sites are indicated by open arrowheads. The enlarged region at the bottom of the figure shows the apparent masses of the confirmed 3CLpro and p1a-22 proteins and the location of their cleavage sites (filled boxes and arrows). The calculated masses of other predicted proteins at the end of ORF 1b are shown as open boxes and arrows (18–20). POL, RNA-dependent RNA polymerase; HEL, helicase.