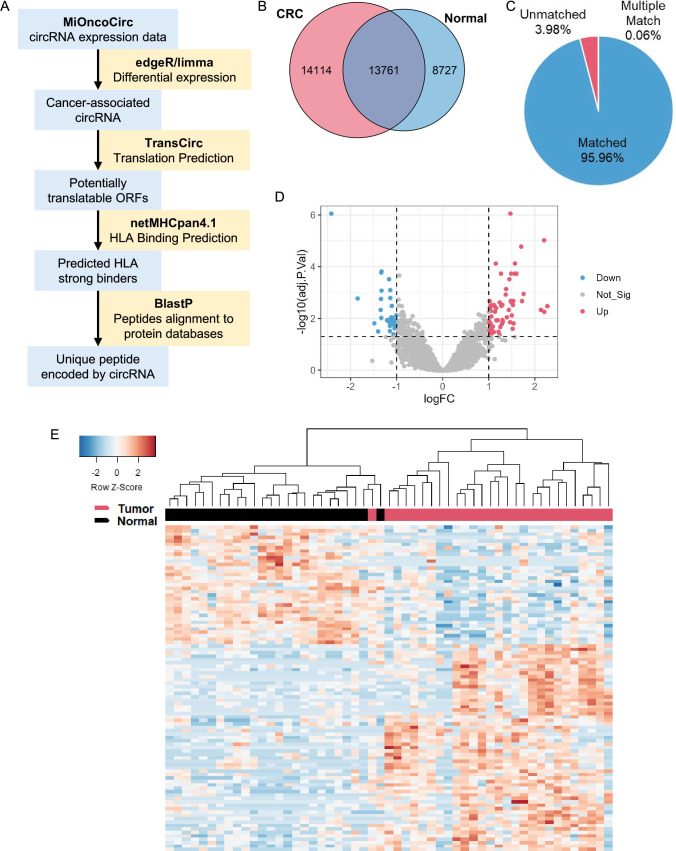
Figure 1.
circRNA expression in MiOncoCirc CRC samples. (A) circRNA neoantigen prediction pipeline. circRNA expression data were acquired from MiOncoCirc and differentially expressed circRNA were analyzed using R package edgeR. circRNA were then mapped to TransCirc database for ORF and translation prediction. HLA-A*11:01 binding affinity of potentially translatable ORFs were predicted using netMHCpan4.1 and shortlisted strong binders were aligned to NCBI protein database using BlastP to screen for novel peptides. (B) circRNA detected in CRC and normal samples. Only overlapped circRNA were kept for downstream analyses (C) fraction of circRNA mapped to TransCirc. The genomic locations of circRNA were used as identifiers to map to TransCirc database. Only uniquely matched circRNA were kept. (D) Volcano plot of differentially expressed circRNA. (E) Expression of differentially expressed circRNA in tumor and normal samples. circRNA, circular RNA; CRC, colorectal cancer; HLA, human leukocyte antigen; ORF, open reading frame.