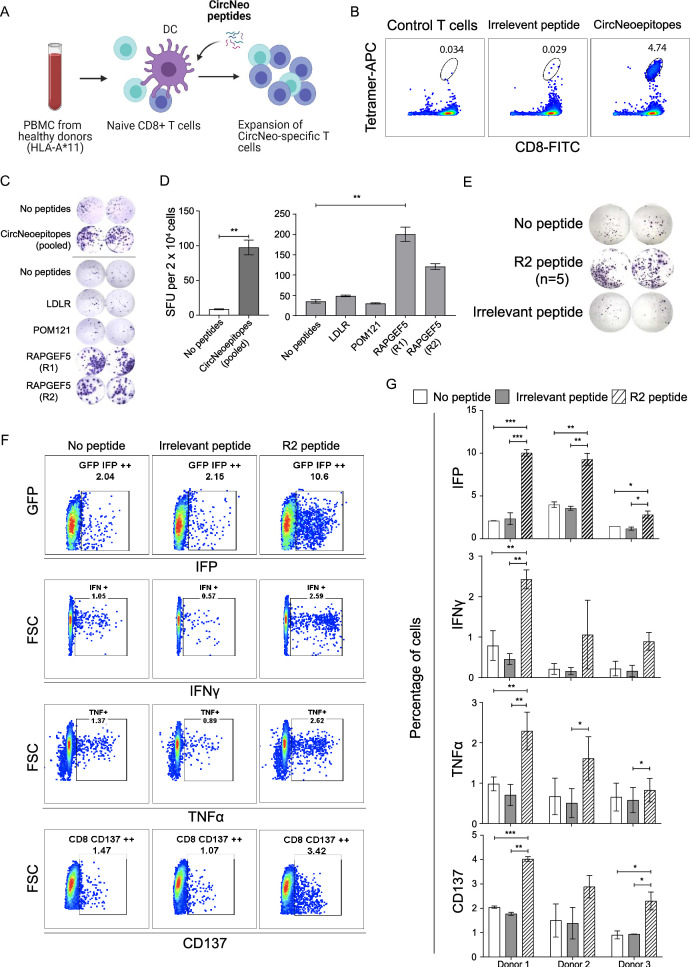
Figure 3.
Immunogenicity validation of shortlisted circRNA neoantigens predicted from MiOncoCirc database. (A) Schematics depicting the flow of training HLA-A*11+ naïve CD8+ T cells and the subsequent immunogenicity validation experiments. Created with BioRender.com. (B) Pooled tetramer and CD8 staining of the circRNA neoantigens specific T cells. (C) ELISpot characterization of IFN-γ secretion by circRNA neoantigens specific T cells after stimulation with HLA-A*11:01 expressing aAPCs, in the absence or presence of pooled peptides and as well as individual peptides encoding the circRNA neoantigens. Images were taken and spots were counted using the CTL ImmunoSpot analyzer. (D) Quantification of the number of IFN-γ spots per 20,000 CD8+ T cells in. Two-tailed unpaired Student’s t-test was used when comparing two experimental groups (pooled ELISpot) and one-way ANOVA was used when comparing five experimental groups (individual ELISpot) **p<0.01. (E) ELISpot quantification of IFN-γ secretion by RAPGEF5_2 (R2) neoantigen peptides specific T cells after stimulation with HLA-A*11:01 expressing aAPCs, in the absence (top panel) or presence (middle panel) of R2 peptide or the irrelevant EBV peptide (bottom panel). (F) Representative FACS plots of granzyme B reporter validation (first row), ICS of IFN-γ (second row), ICS of TNF-α (third row) and expression of cluster of differentiation 137 (fourth row) in R2 specific T cells in the absence of peptides (first column), presence of irrelevant EBV peptides (second column) and the presence of R2 peptide (third column). (G) Quantification of the representative FACS plots in (F) for trained T cells from three different independent healthy donors. n=3 biological samples per group, each experiment was performed with R2 neoantigen-specific T cells trained independently from three different healthy donors. One-way ANOVA was used for multiple experimental groups with p value adjustment. Data is shown as mean±SEM. *P<0.05; **p<0.01; ***p<0.001. aAPC, artificial antigen-presenting cells; ANOVA, analysis of variance; circRNA, circular RNA; DC, dendritic cell; EBV, Epstein-Barr virus; ELISpot, enzyme-linked immunospot; GFP, green fluorescent protein; HLA, human leukocyte antigen; ICS, intracellular cytokine staining; IFN, interferon; IFP, infrared fluorescent protein; PBMC, peripheral blood mononuclear cell; SFU, spot-forming units; TNF, tumor necrosis factor; FSC, forward scatter; FACS, fluorescence-activated cell sorting; FITC, fluorescein isothiocyanate.