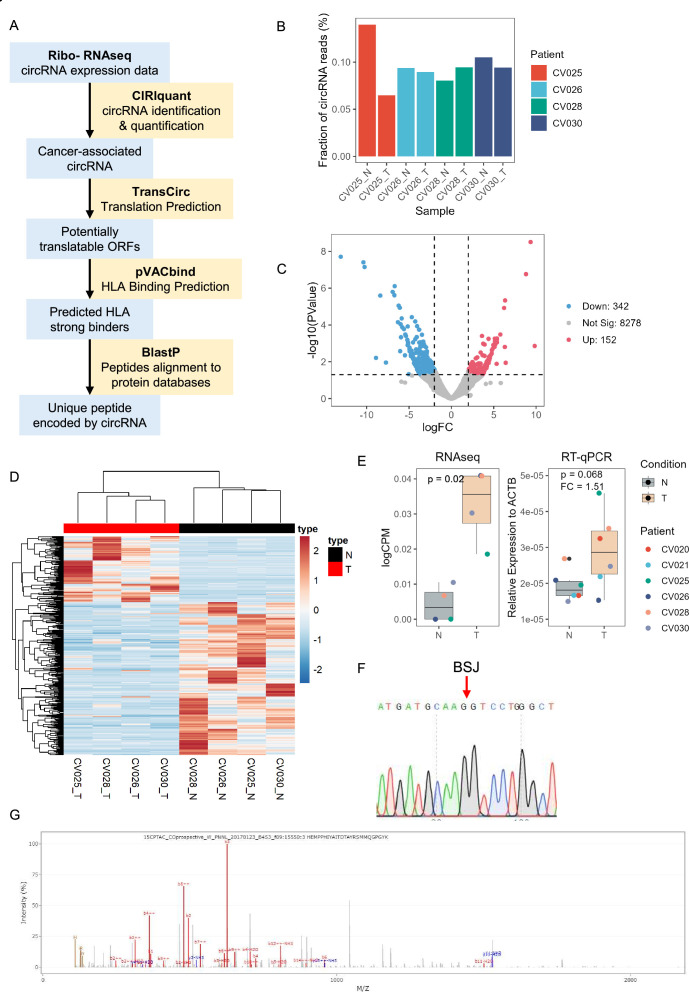
Figure 4.
circRNA expression in paired tumor and normal samples from patients with CRC. (A) circRNA neoantigen prediction pipeline. RNA from three pairs of tumor and normal samples were sent for ribosome-depletion RNA-seq. circRNA were identified and quantified using CIRIquant. circRNA were then mapped to TransCirc database for ORF and translation prediction. HLA binding affinity of potentially translatable ORFs were predicted using pVACbind, which employed four algorithms for HLA binding affinity prediction, to increase the sensitivity and specificity. Shortlisted strong binders were aligned to NCBI protein database using BlastP to screen for novel peptides. (B) Fraction of total mapped RNA and circRNA identified from each sample (n=1). (C) Volcano plot of differentially expressed circRNA. (D) Expression of differentially expressed circRNAs in tumor and normal samples from RNA-seq data. (E) Expression of circMYH9. Left, expression data from RNA-seq. Right, Relative expression in six pairs of tumor and normal samples from patients with CRC by RT-qPCR. (F) Sanger sequencing result showing the back splicing junction of circMYH9 from RT-qPCR amplicon. (G) Representative PSM of circMYH9 peptide SMMQGPGYK. Empirical Bayes moderated t-statistics from edgeR and the paired two-sided Student’s t-test were used in (E) for RNA-seq and RT-qPCR, respectively. The boxes in boxplots represented the median ±1 quartile, with the whiskers extending to the largest or smallest non-outlier value within 1.5 times the IQR from the third quartile and first quartile, respectively. BSJ, back-splicing junction; circRNA, circular RNA; CRC, colorectal cancer; HLA, human leukocyte antigen; ORF, open reading frame; RNA-seq, RNA sequencing; RT-qPCR, quantitative real-time PCR; CPM, counts per million; ACTB, actin beta.