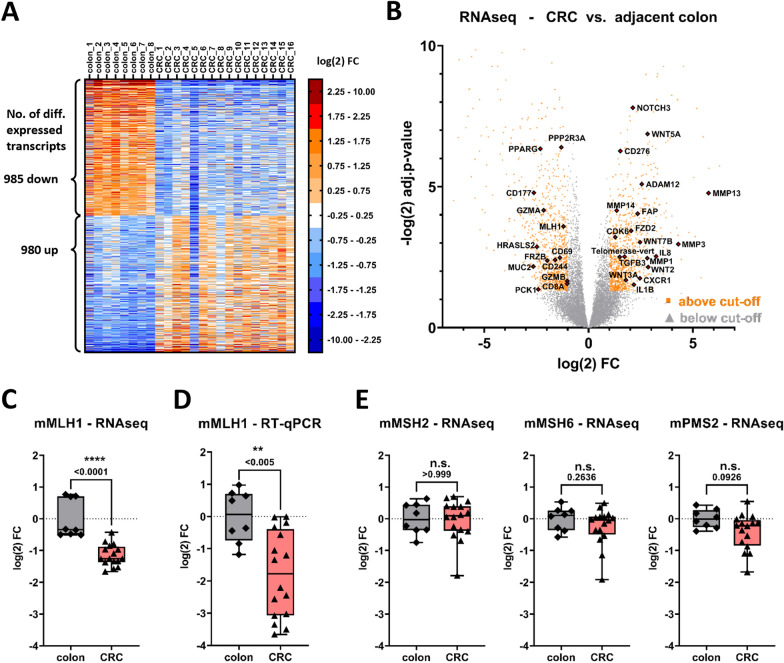
Fig. 4.
Transcriptomics reveals similar DEG signatures in rhesus and human CRCs and indicates transcriptional downregulation of MLH1. A RNAseq revealed 1965 significantly differentially expressed genes (985 up, 980 down) in rhesus CRC compared to adjacent healthy colon. B Genes frequently dysregulated in human cancers are similarly altered in rhesus CRC including upregulation of MMPs, FAP, telomerase, tumor-promoting or associated pathways and reduction of GZMA, FRZB, CD69, and PPARG. A statistically significant downregulation of MLH1 was determined in rhesus CRC by (C) RNAseq and corroborated by (D) RT-qPCR. E In contrast, no difference was observed for the other MMR proteins MSH2, MSH6, and even PMS2. Statistical analysis was performed by Mann–Whitney U test and a p < 0.05 was considered statistically significant