FIGURE 1.
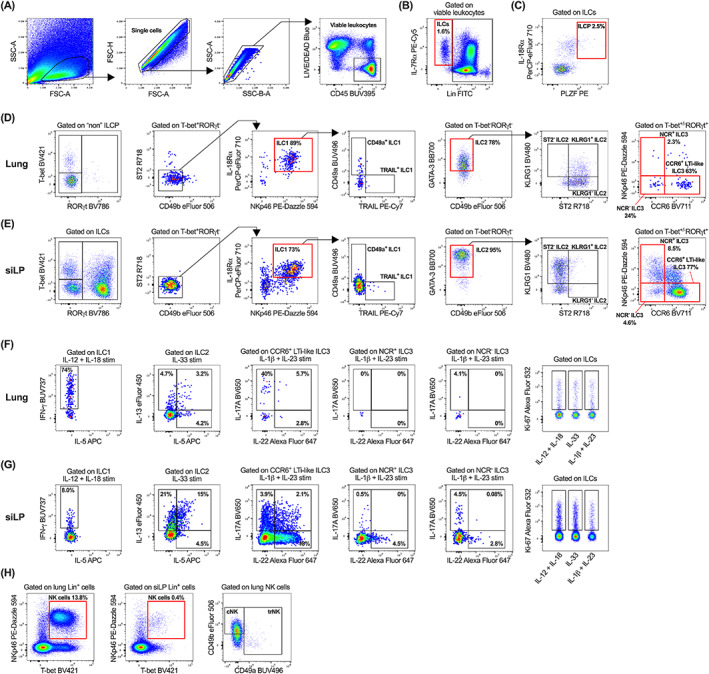
Gating strategy to characterize innate lymphoid cell (ILC) populations and their subset‐specific cytokine and proliferative profiles. After exclusion of cellular debris, doublets, red blood cell contamination and non‐viable CD45+/− cells, viable CD45+ leukocytes (A) were gated on IL‐7Rα and lineage (CD3, CD4, CD5, CD11b, CD11c, CD19, B220, F4/80, FcεR1, Gr1, TCRβ, TCRγδ, Ter119; Lin) negative cells to identify ILCs (B). ILCPs were then identified as IL‐18Rα+PLZF+ cells (C). Characterization of ILC1, ILC2, NCR+, NCR− and CCR6+ LTi‐like ILC3 subsets in the lung (D) and siLP (E). All samples in panel (A–E) were ex vivo stimulated for 5 h with PMA/Ionomycin/BFA. Single cell suspensions were ex vivo stimulated for 5 h with PMA/Ionomycin/BFA and either IL‐12 and IL‐18 (pro‐ILC1), IL‐33 (pro‐ILC2), or IL‐1β and IL‐23 (pro‐ILC3) and ILC subsets were assessed for production of subset‐specific cytokines (IFN‐γ, IL‐5, IL‐13, IL‐17A, and IL‐22) and cellular proliferation (Ki‐67) within the lung (F) and siLP (G). (H) Identification of NK cells within the lung and siLP, with downstream characterization of lung tissue‐resident NK (trNK) and circulating NK (cNK) cells. Plots in panel (A–C) are representative of individual lung samples. For panels (B–H), viable leukocytes from three technical lung or siLP replicates were concatenated into single FCS files prior to gating. All fluorochrome‐conjugated antibodies were titrated (Figure S1) during panel optimization and manual gating was determined using fluorescence minus one controls (Figure S2) where necessary [Color figure can be viewed at wileyonlinelibrary.com]
