Extended Data Fig. 9 |. Overview of the iCat2/GAIN Study.
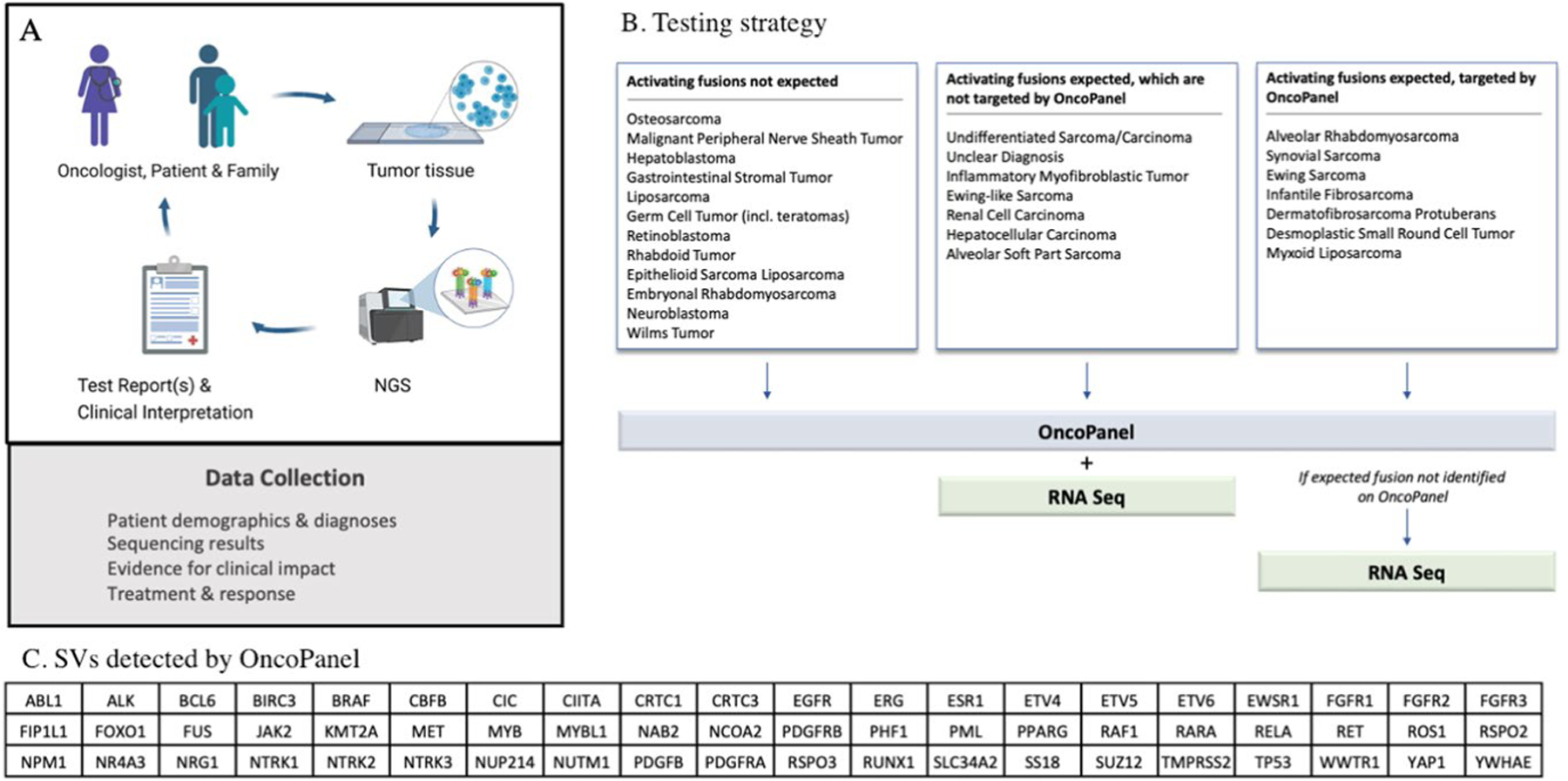
Overview of the iCat2/GAIN study (a, created with Biorender.com). Targeted DNA NGS is performed on one or more tumor samples from each patient. Selected patients also have tumors subjected to RNA sequencing. Test results are returned to the treating oncologist and follow-up treatment and response data are collected. Details of clinical interpretation of test reports including molecular tumor board are shown in extended data Fig. 3. Testing strategy (b) to select patients for additional sequencing with either whole transcriptome sequencing or targeted RNA fusion panel testing (RNASeq). RNASeq was not performed if it was unlikely to contribute to research findings or clinical care. In this study, transcriptome sequencing was analyzed only for structural variants (SVs) and OncoPanel detects rearrangements in 60 genes (c). The testing triage is based on several assumptions: 1) False positives for SV detection OncoPanel are uncommon; 2) If oncogenic fusions have not been described in a particular solid tumor in previous studies and typical oncogenic events for that diagnosis are present then novel oncogenic fusions are unlikely; and 3) very rare pediatric solid malignancies might harbor previously undescribed fusions because they are understudied.
