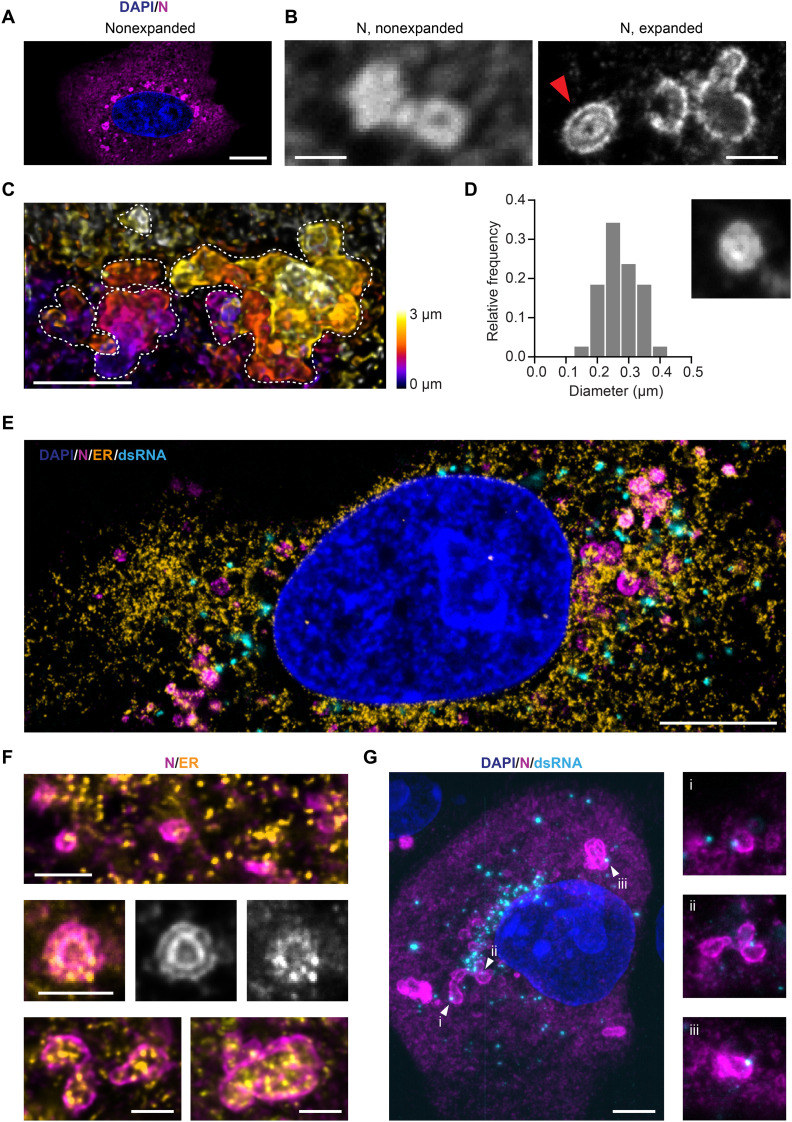
Fig. 3. The SARS-CoV-2 N protein is organized in layered structures that are strongly interwoven with the topology of the ER and contain RNA replication foci.
(A) Confocal microscopy cannot resolve the substructure of N protein puncta in the cytosol of infected Vero cells (blue: nuclei; magenta: N protein). Scale bar, 10 μm. (B) The combination with expansion microscopy reveals double layers of N protein within the larger compartments (red arrow). Scale bars, 1 μm. Linear expansion factor of 4.2. (C) Combining expansion with light sheet microscopy reveals the convoluted nature of the N protein structures in 3D. Dotted lines outline the boundaries of each convoluted compartment in the maximum intensity projection. Scale bar, 2 μm. Expansion factor of 4.2. (D) The inner circular layer of the N protein double-layer compartments has an average diameter of 275 nm. Thirty-eight compartments from 10 cells at 12 hpi were analyzed. The micrograph size is 1.25 μm per side. (E) Representative confocal image of an expanded, infected Vero cell stained for the nucleus (blue), ER (calnexin; orange), N protein (magenta), and dsRNA (cyan). Scale bar, 5 μm. Expansion factor of 4.2. (F) The N protein (magenta) forms layers around ER membranes (yellow). Scale bars, 1 μm. Expansion factor of 4.2. (G) A combination of expansion and light sheet microscopy reveals that dsRNA foci (cyan) sit in the layers of the N compartments (magenta). Scale bar, 5 μm. Expansion factor of 4.2. The size of the smaller micrographs is 5 μm per side.