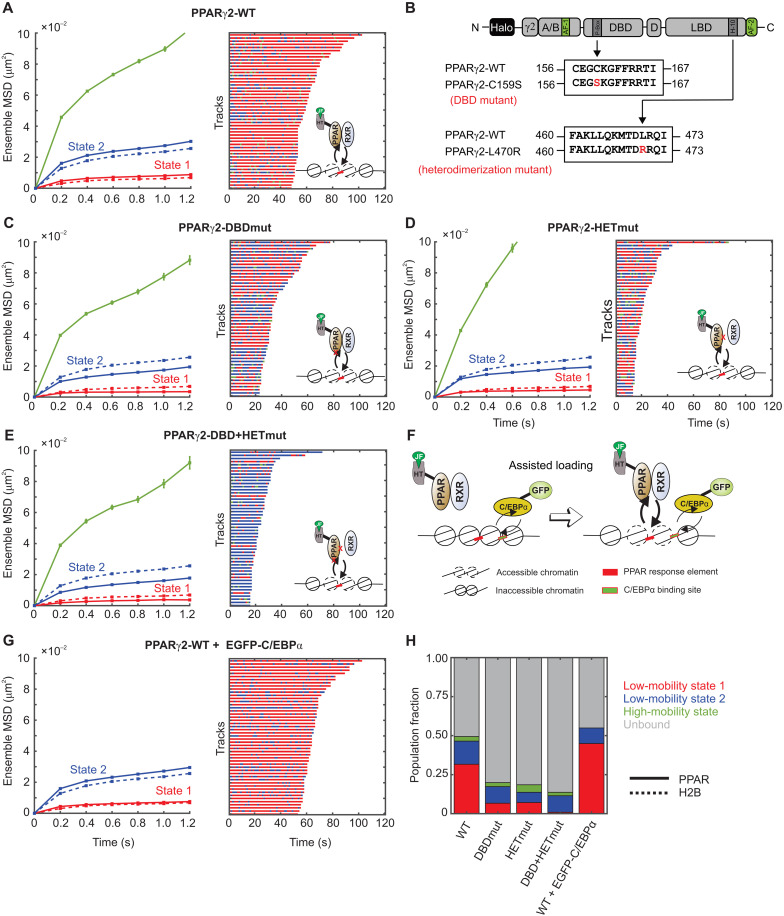
Fig. 5. State 1 for PPARγ2 requires intact DBD and the ability to form heterodimeric complexes.
(A) Left: Ensemble MSD of H2B (dashed lines, Ncell = 54, Ntracks = 8522, and Nsub-tracks = 29,262) and wild-type PPARγ2 (solid lines, Ncells = 127, Ntracks = 20,983, and Nsub-tracks = 62,848). Error bars indicate SEMs. Right: Temporal reconstruction of the 50 longest tracks along with (inset) a cartoon depicting PPARγ2 binding to PPAR response elements (PPRE). (B) Schematic of point mutations to abrogate the DBD and heterodimerization domains of PPARγ2. (C to E) Left: Ensemble MSD for indicated PPARγ2 mutant. Error bars denote SEMs. Right: Temporal reconstruction of the 50 longest tracks colored by state assignment: (C) PPARγ2-DBD mutant (PPARγ2-DBDmut) (Ncells = 38, Ntracks = 3721, and Nsub-tracks = 9872), (D) PPARγ2-heterodimerization mutant (PPARγ2-HETmut) (Ncells = 28, Ntracks = 1728, and Nsub-tracks = 4049), and (E) PPARγ2-DBD + HET mutant (Ncells = 46, Ntracks = 1695, and Nsub-tracks = 4046). (F) Assisted loading model for C/EBPα-mediated PPARγ2 loading. (G) Left: Ensemble MSD for PPARγ2-WT with overexpression of GFP-C/EBPα (Ncells = 89, Ntracks = 18,912, and Nsub-tracks = 63,842). Error bars denote SEMs. Right: Temporal reconstruction of the 50 longest tracks. (H) Comparative population fractions for all PPARγ2 variants.