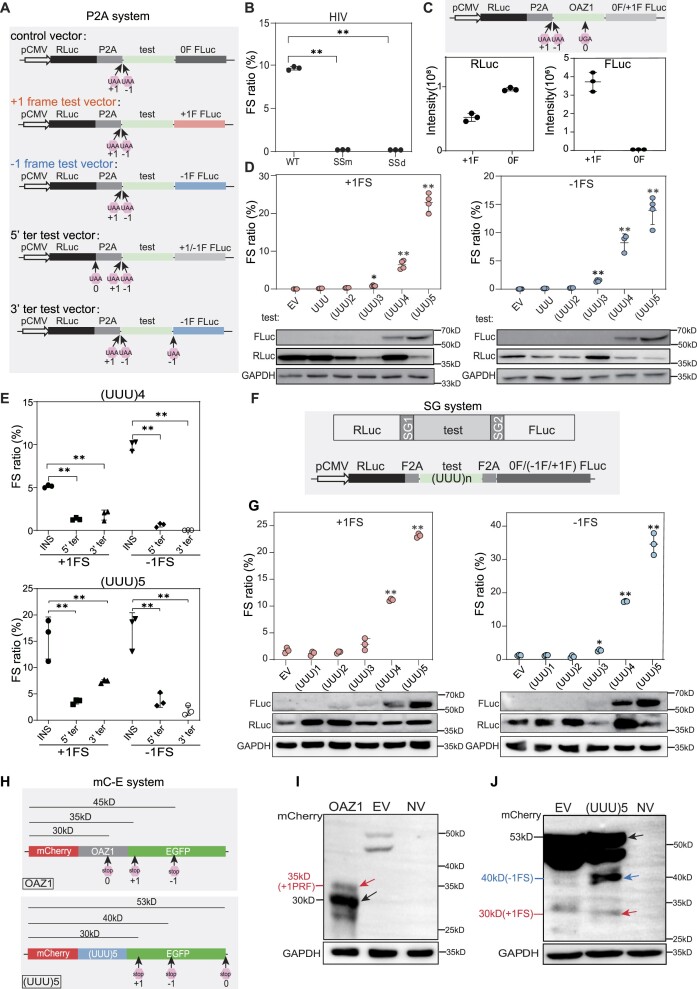
Figure 1.
UUU codon repeats could trigger robust ribosomal frameshifting. (A) Schematic diagram of the P2A dual luciferase system (RLuc, Renilla luciferase; FLuc, firefly luciferase). 0F, +1F or −1F FLuc indicate the FLuc sequence is placed in the 0, +1 or −1 frame, respectively. 0, +1 and −1 UAA mean that a UAA stop codon was added in the 0, +1 or −1 frame, respectively. (B) Bar plot showing frameshifting ratio of HIV −1 PRF sequence in HEK293T cells by luciferase assays. SSm, slippery site mutation (UUUUUUA to CUUCUUA). SSd, slippery site deletion. (C) Bar plot showing Renilla luciferase (left) and firefly luciferase (right) activity of OAZ1 sequences. +1F and 0F means construct with FLuc in the +1 or 0 frame, as shown in (A) The cartoon on top showed the reporter structure. (D) Detection of ribosomal frameshifting induced by different number of UUU codon repeats with dual luciferase systems (top panel) and western blots (bottom panel). EV, empty vector. (E) Frameshifting ratio induced by (UUU)4 or (UUU)5 insertion (INS) with different vectors shown in (A). +1FLuc and −1FLuc means construct with FLuc in the +1 or −1 frame. (F) Schematic diagram of Stop-Go (SG) system. SG1 and SG2 indicate two F2A sequence. (G) Frameshifting ratios (top) and western blot results (bottom) from SG system with different number of UUU repeat insertion in HEK293T cells. EV, empty vector. (H) Schematic diagram of constructs (mC-E system) with OAZ1 (top) and (UUU)5 (bottom) insertion. 0, +1 and −1 mean the position of first stop codon in the corresponding frame after mCherry coding sequence. (I) Western blot analysis for the frameshifting proteins induced by OAZ1 sequence. NV, transfection without vector. (J) Western blot analysis the frameshifting induced by (UUU)5 codon in HEK293T cells. In this figure, FS stands for ribosomal frameshifting, EV for empty vector, error bars for standard deviation of three biological replicates. (*) P< 0.05, (**) P < 0.01 (Student's t-test for 2 data sets and one-way ANOVA for >2 data sets).