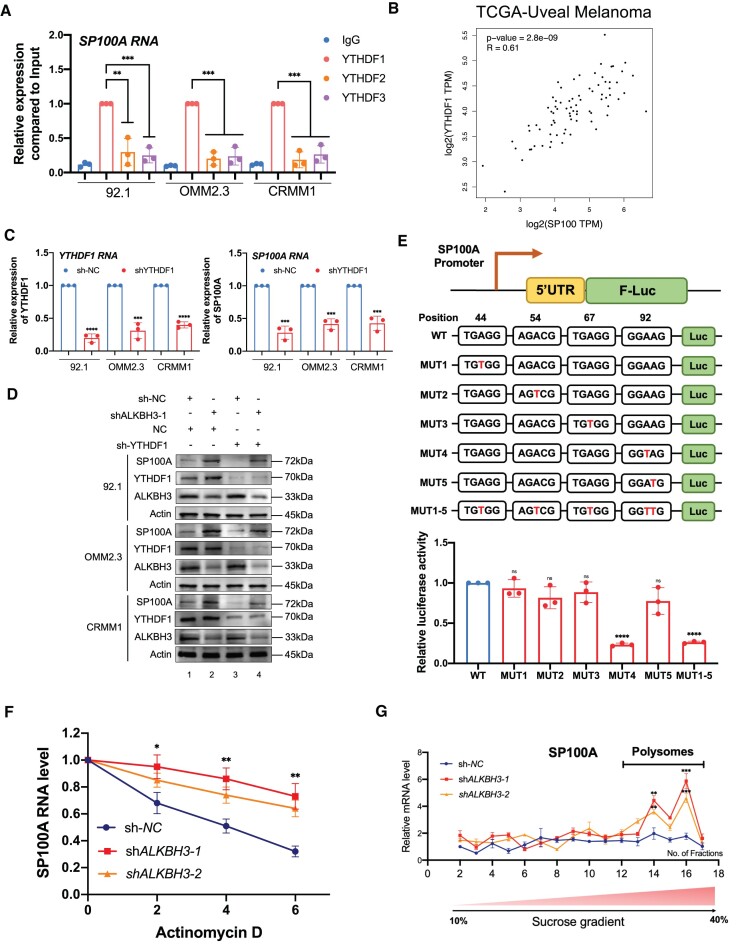
Figure 7.
m1A modification of SP100A increases its RNA stability and translational efficacy. (A) RIP-qPCR assay of SP100A expression in ocular melanoma cells (92.1, OMM2.3 and CRMM1) by YTHDF1, YTHDF2 and YTHDF3. The data are presented as the mean ± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. **P < 0.01, ***P < 0.001. (B) Correlation analysis of SP100A expression and YTHDF1 expression in the TCGA-UM cohort (n = 80). Significance was determined by Pearson correlation analysis (R = 0.61, P < 0.0001). (C) qPCR data showing YTHDF1 RNA expression and SP100A RNA in ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon YTHDF1 knockdown. The data are presented as the mean ± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. ***P < 0.001, ****P< 0.0001. (D) Western blot showing YTHDF1, SP100A, ALKBH3 expression relative to ACTB expression in ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon ALKBH3 and YTHDF1 knockdown. (E) The luciferase reporter gene assay demonstrated the relative luciferase activity of the wild-type and six mutant SP100A 5′UTR reporter vectors. The data are presented as the means ± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. ****P < 0.0001. (F) Half-life of SP100A in wild-type and ALKBH3-deficient 92.1 cells treated with actinomycin (5 g/ml) for 0–6 h. (G) Polysome profiling assays of 92.1 cells with or without ALKBH3 knockdown. RNAs in different ribosome fractions were extracted and subjected to qPCR analysis. Data are shown as the mean ± SD. Significance was determined by unpaired two-tailed Student's t test. ***P < 0.001, ****P < 0.0001.