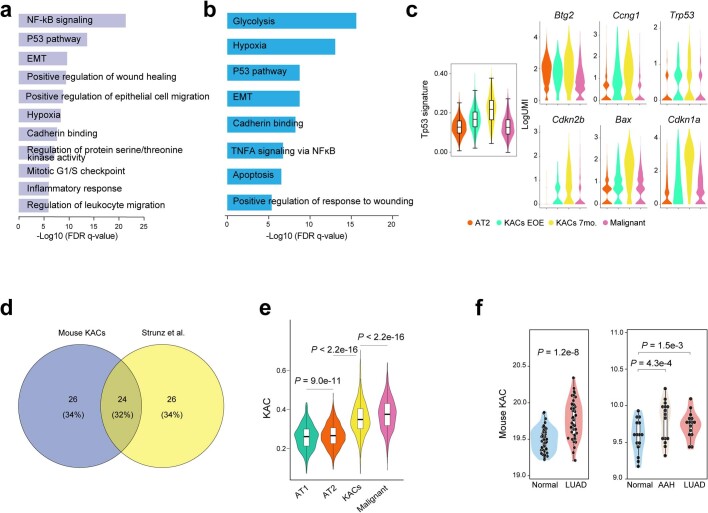
Extended Data Fig. 9. Mouse KAC signatures and pathways are relevant to both injury models and human KM-LUAD.
a,b, Pathway enrichment analysis of KACs relative to other alveolar cell subsets and malignant cells in tumour-bearing mice at 7 months following NNK (a) and in the human LUAD scRNA-seq dataset from this study (b). c, Enrichment of Tp53 signature derived from mouse KACs, and expression of Btg2, Ccng1, Cdkn2b, Bax, Cdkn1a, as well as Trp53 itself, across AT2 cells, malignant cells, and KACs at EOE or at 7 months following NNK or saline. n cells in each group: AT2 = 1,791; KACs EOE = 301; KACs 7mo. = 335; Malignant =1,693. d, Pie chart showing percentages of unique and overlapping DEG sets between mouse KACs from this study and Krt8+ transitional cells identified by Strunz and colleagues. e,f, Expression of the mouse KAC signature across alveolar and malignant cell subsets from this study (e), in normal lung (Normal) and LUAD tissues from the TCGA cohort (f, left), as well as in normal lung (Normal), AAH, and LUAD tissues of our premalignancy cohort (f, right). n cells in each group of panel e: AT2 = 1,791; KACs EOE = 301; KACs 7mo. = 335; Malignant = 1,693. n samples in each group of panel f left: Normal = 52; LUAD = 52. n samples in each group of panel f right: Normal = 15; AAH = 15; LUAD = 15. Box-and-whisker definitions are similar to Extended Data Fig. 1f. P values were calculated using two-sided Wilcoxon Rank-Sum test with a Benjamini–Hochberg correction.