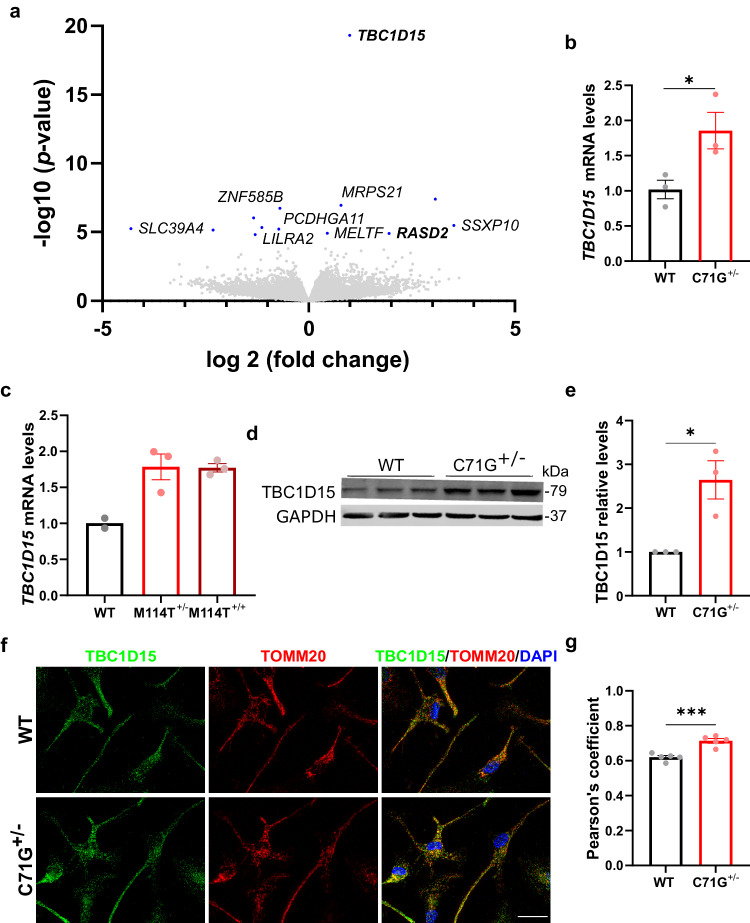
Fig. 3. TBC1D15 is upregulated in ALS-PFN1 iMGs and colocalizes with mitochondria.
a Volcano plot of differentially expressed genes between ALS-PFN1 iMGs (n = 4 for C71G+/− iMGs and n = 3 for M114T+/− iMGs) relative to WT iMGs (n = 7), where n refers to an independent differentiation; see Data S5. Differentially expressed genes (P-adjusted value < 0.05) are highlighted in blue. b, c Relative mRNA levels of TBC1D15 normalized to the average of the respective WT iMGs. b *P = 0.0452, t = 2.875, df = 4 for n = 3 independent differentiations; c WT n = 2, M114T+/− and M114T+/+ n = 3 independent differentiations. d TBC1D15 protein expression determined by Western blot analysis. e Quantification of (d). TBC1D15 levels were normalized to GAPDH and then to the levels of the respective WT line from the same differentiation (*P = 0.0197, t = 3.763, df = 4) for n = 3 independent differentiations. f, g Colocalization analysis of TBC1D15 and TOMM20 in WT and C71G+/− iMGs. f Representative immunofluorescence images of TBC1D15 (green), TOMM20 (red), and merged images including DAPI (blue) showing regions of overlayed TBC1D15:TOMM20 signal in yellow. Scale bar = 25 µm. g Pearson’s correlation coefficient of TBC1D15 and TOMM20 signal (***P = 0.0004, t = 5.714, df = 8) for n = 5 independent differentiations. All bar graphs show mean ± SEM, where each data point represents an independent differentiation. Statistics were determined using unpaired two-tailed t-test for WT vs C71G+/− iMGs comparisons or ordinary one-way ANOVA with Dunnett’s multiple comparisons test for WT vs M114T+/− and M114T+/+ comparisons. Source data are provided as a Source Data file.