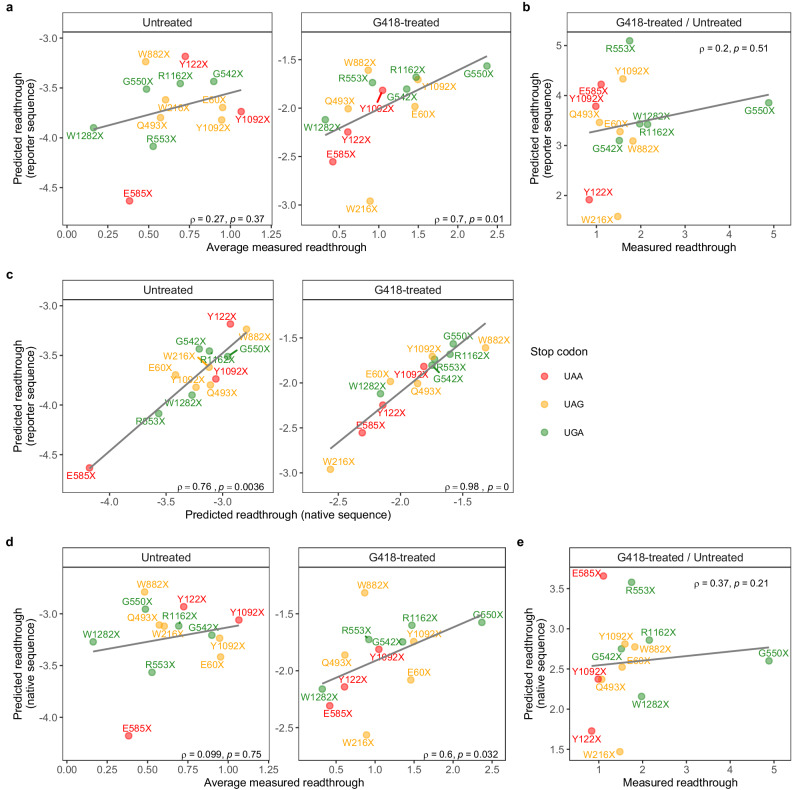
Fig. 4. Random forest model can accurately predict the readthrough of CFTR PTC alleles in G418-treated cells.
a Readthrough measured by Dual-Luc assay (average of 4–7 replicates) vs. readthrough predicted by random forest model using CFTR PTC alleles in Dual-Luc reporter’s sequence. b Response to G418 treatment is defined as fold-change of readthrough in G418-treated to untreated condition from (a), measured vs. predicted. c Comparison of two readthrough prediction schemes: predicted using CFTR PTC allele in reporter’s sequence or predicted using CFTR PTC allele’s native sequence. d As in (a), but readthrough was predicted using CFTR PTC allele’s native sequence. e As in (b), but readthrough was predicted using CFTR PTC allele’s native sequence. For all panels, the two-tailed Spearman’s correlation coefficient (ρ) and the associated p-value is reported. Source data are provided as a Source Data file and Supplementary Data 1.