FIGURE 1.
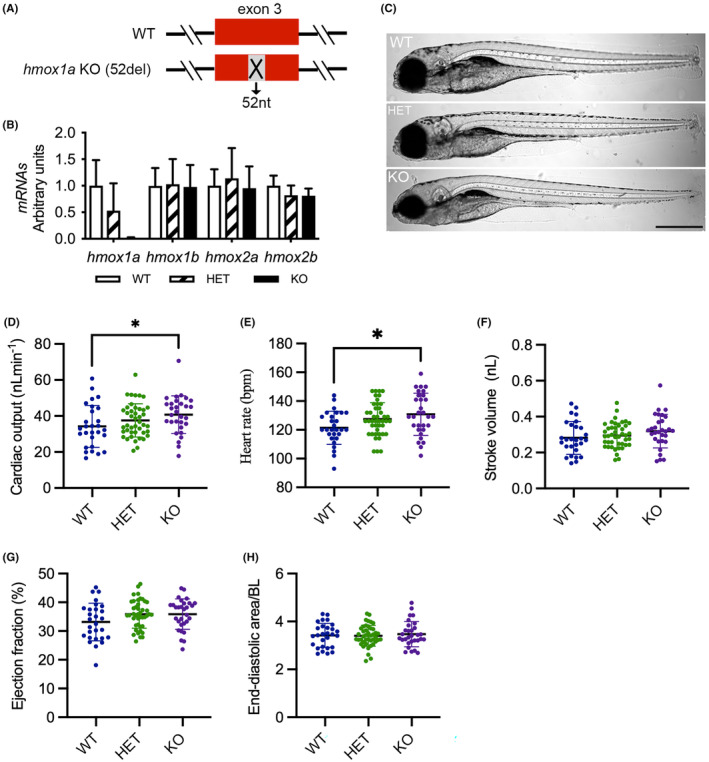
Deletion of hmox1a increases cardiac output in larvae. (A) Scheme of CRISPR/Cas9‐generated 52‐base pair deletion in exon 3 of hmox1a. (B) RT‐qPCR analysis of hmox1a, hmox1b, hmox2a and hmox2b in wild type (WT), heterozygous (HET) and homozygous (KO) hmox1a mutant at 5–6 dpf. The results are presented as fold change compared to WT. RNA was extracted from 5 to 10 pooled larvae. The number of RNA samples used for RT‐qPCR analyses as following: WT n = 5; HET(52del) n = 6; KO(52del) n = 6. (C) Representative images of hmox1a WT, HET(52del) and KO(52del) larvae at 5 dpf showing no obvious gross phenotype. Scale bar: 500 μm. (D–H) Analysis of cardiac function at 5–6 dpf larvae indicating an increase in cardiac output (D) and heart rate (E), and no significant change in stroke volume (F), ejection fraction (G) and end‐diastolic ventricular area normalized to body length (H) in hmox1a KO(52del). WT n = 28; HET(52del) n = 43; KO(52del) n = 30. Data are presented as mean ± SD. One‐way ANOVA with Tukey adjustment for multiple comparisons. *p < 0.05.
