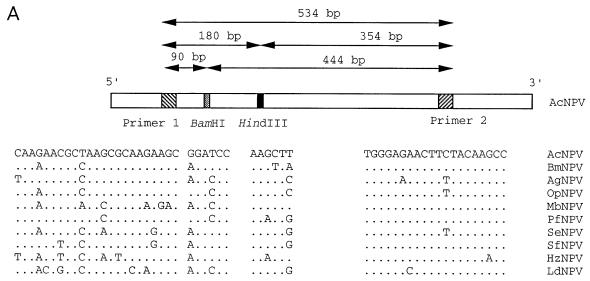
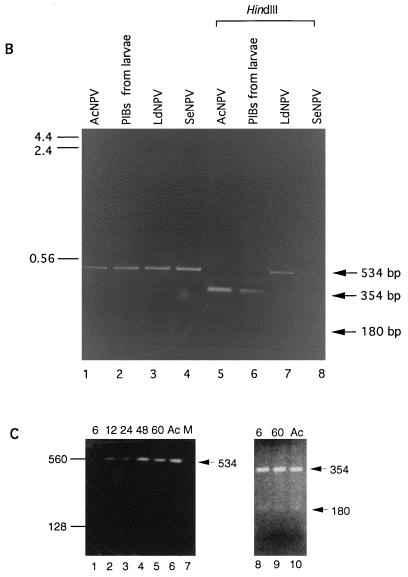
FIG. 1.
(A) Sequence comparisons of various baculovirus polyhedrin gene sequences in the region used for PCR primers and restriction analysis. Abbreviations: Bm, Bombyx mori; Ag, Anticarsia gemmatalis; Op, Orgyia pseudotsugata; Mb, M. brassicae; Pf, Panolis flammea; Se, S. exigua; Sf, S. frugiperda; Hz, H. zea. Primer pairs F (5′-AGAACGCTAAGCGCAAGAAGCA-3′) and R (5′-GGCTTGTAGAAGTTCTCCCA-3′) are based on the AcNPV polyhedrin sequence and map to nucleotides 4603 to 4625 and 5136 to 5117 of the AcNPV genome, respectively (1). Locations of diagnostic HindIII and BamHI sites and sizes of predicted PCR product and restriction fragments are indicated. (For the polh sequence, see references 3 and 37). (B) PCR products amplified by using primer pairs specific to the AcNPV polyhedrin gene. AcNPV DNA (lane 1), DNA isolated from OV from moribund AcNPV-inoculated L. dispar larvae (lane 2), LdNPV DNA (lane 3), and S. exigua NPV DNA (lane 4) were amplified. The amplified products from lanes 1 to 4 were digested with HindIII (lanes 5 to 8, respectively). Predicted amplification and digestion products are indicated by arrows to the right of the panel. The size markers on the left are given in kilobase pairs. (C) PCR products amplified by using the AcNPV polh primer pair from DNA isolated from third-instar L. dispar midgut tissues or AcNPV DNA as the template. AcNPV-infected larvae at 6, 12, 24, 48, and 60 h p.i. (lanes 1 to 5, respectively), AcNPV DNA (lane 6), or mock-infected larvae (lane 7) were used. Amplified products from 6 and 60 h p.i. were digested with HindIII (lanes 8 and 9, respectively) and compared to the HindIII-digested amplification product from AcNPV DNA (lane 10). The predicted 354- and 180-bp restriction fragments are indicated with arrows. Size markers on the left are given in base pairs.