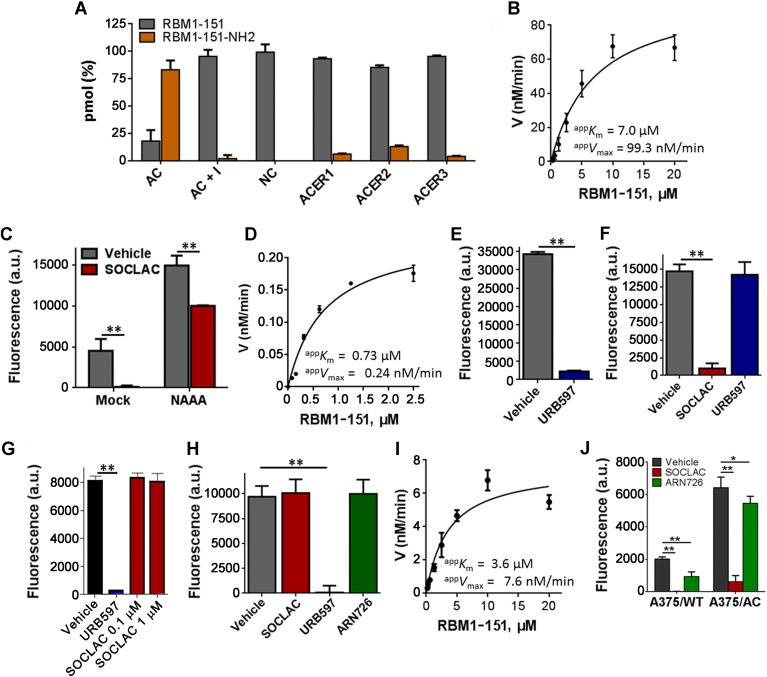
Fig. 1.
Hydrolysis of RBM1-151 in cell-free systems. A: RBM1-151 (20 μM) was incubated for 1 h in the appropriate buffer with recombinant NC (5 ng), lysates of A375/AC cells (20 μg) (AC) or ASAH2-null MEFs (140 μg) (ACER3) or microsomes (100 μg) from HeLa TRex ACER1 (ACER1) or HeLa TRex ACER2 cells (ACER2) induced with tetracycline, and the lipid extracts were analyzed by UPLC-HRMS. SOCLAC (I) (1 μM) was added to the AC375/AC lysate (AC + I) 1 h prior substrate addition. B: Hydrolysis of different concentrations of RBM1-151 by A375/AC cell lysates (20 μg) in acid buffer. Incubation time was 30 min. Michaelis-Menten analysis gave appKm = 7.0 μM; appVmax = 99.3 nM/min. C: Lysates of HEK293/NAAA or HEK293/mock (10 μg protein) were incubated for 3 h with RBM1-151 (5 μM) at acid pH in the presence or the absence (vehicle) of SOCLAC (1 μM), and the reaction mixture was processed for fluorescence release as detailed in the Materials and methods section. D: Hydrolysis of different concentrations of RBM1-151 by SOCLAC-pretreated HEK293/NAAA cell lysates (5 μg). Incubation time was 3 h at acid pH. Michaelis-Menten analysis gave appKm = 0.73 μM; appVmax = 0.24 nM/min. E: Lysates (25 μg protein) of LNCaP cells were incubated for 3 h with RBM1-151 (10 μM) in neutral buffer B in the presence or the absence (vehicle) of URB597 (50 μM), and the reaction mixture was then processed for fluorescence release as detailed in the Materials and methods section. F: Lysates (25 μg protein) of LNCaP cells were incubated for 3 h with RBM1-151 (10 μM) at acid pH in the presence or the absence (vehicle) of URB597 (50 μM) or SOCLAC (0.1 μM), and the reaction mixture was then processed for fluorescence release as detailed in the Materials and methods section. G: Microsomes (50 μg protein) from LNCaP cells were incubated for 3 h with RBM1-151 (10 μM) in neutral buffer B in the presence or the absence (vehicle) of URB597 (50 μM) or SOCLAC, and the reaction mixture was then processed for fluorescence release as detailed in the Materials and methods section. H: Lysates (25 μg protein) of ASAH2-null MEFs were incubated for 3 h with RBM1-151 (10 μM) in neutral buffer B in the presence or the absence (vehicle) of URB597 (50 μM), SOCLAC (0.1 μM), or ARN726 (0.1 μM), and the reaction mixture was then processed for fluorescence release as detailed in the Materials and methods section. I: Hydrolysis of different concentrations of RBM1-151 by ASAH2-null MEF lysates (20 μg). Incubation time was 30 min. Michaelis-Menten analysis gave appKm = 3.6 μM; appVmax = 7.6 nM/min. J: Lysates of A375/WT and A375/AC cells (20 μg) were incubated for 3 h with RBM1-151 (10 μM) in acid buffer in the presence or the absence (vehicle) of SOCLAC (0.1 μM) or ARN726 (0.1 μM), and the reaction mixture was then processed for fluorescence release as detailed in the Materials and methods section. Data (mean ± SD) were obtained from two (A, C, D, and J) or three (B, E, F, G, and H) different experiments with triplicates. Statistical significance between means was analyzed by one-way ANOVA followed by Tukey multiple comparison test. Asterisks indicate statistical significance at ∗P ≤ 0.001 and ∗∗P ≤ 0.0001. The enzyme sources and buffers used for each enzyme are summarized in supplemental Table S1. That SOCLAC and SACLAC have almost identical activity as AC inhibitors is shown in supplemental Fig. S1. AC, acid ceramidase; MEF, mouse embryonic fibroblast; NAAA, N-acylethanolamine-hydrolyzing acid amidase; NC, neutral ceramidase; SACLAC, (2S,3R)2-chloro-N-(1,3-dihydroxyoctadecan-2-yl)acetamide; SOCLAC, (2S,3R,E)2-chloro-N-(1,3-dihydroxyoctadec-4-en-2-yl)acetamide.