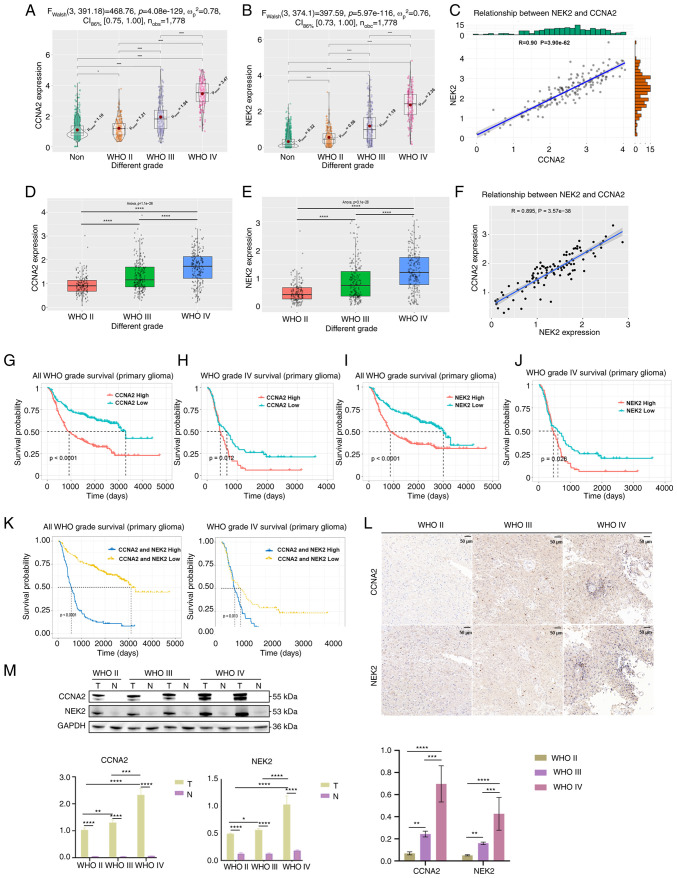
Figure 1.
Highly expressed CCNA2 and NEK2 in glioma. (A) Analysis of NEK2 expression in different WHO grades of glioma, using TCGA database (*P<0.05 and ****P<0.0001, with independent t-test). (B) Analysis of NEK2 expression in different WHO grades of glioma, using TCGA database. (C) Co-expression analysis of CCNA2 and NEK2 expression in GBM, using TCGA database (P<0.0001, with Pearson correlation coefficient). (D) Analysis of NEK2 expression in different WHO grades of glioma, using the CGGA database. (E) Analysis of NEK2 expression in different WHO grades of glioma, using the CGGA database (****P<0.0001, with one-way ANOVA followed by Tukey's multiple comparison test). (F) Co-expression analysis of CCNA2 and NEK2 expression in GBM, using the CGGA database (P<0.0001, with Pearson correlation coefficient). (G) The Kaplan-Meier analysis of overall survival in patients with glioma based on CCNA2 expression, using the CGGA database (P<0.0001, with log-rank test). (H) The Kaplan-Meier analysis of overall survival in patients with GBM based on CCNA2 expression, using the CGGA database (P=0.012, with log-rank test). (I) Kaplan-Meier analysis of overall survival in patients with glioma based on NEK2 expression, using the CGGA database (P<0.0001, with log-rank test). (J) The Kaplan-Meier analysis of overall survival in patients with GBM based on NEK2 expression, using the CGGA database (P=0.026, with log-rank test). (K) The Kaplan-Meier analysis of overall survival in patients with glioma and GBM based on CCNA2 and NEK2 expression, using the CGGA database (P<0.0001, for all glioma and P=0.013, for GBM, respectively, with log-rank test). (L) Immunohistochemical staining of CCNA2 and NEK2 in GBM samples, compared with adjacent normal tissue. (M) Protein expression of CCNA2 and NEK2 in GBM samples, compared with adjacent normal tissue. T, tumor tissue; N, adjacent normal tissue. Original blots are presented in Fig. S1. Western blot and immunohistochemical analyses were conducted to detect CCNA2 and NEK2 protein levels in GBM samples (*P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001, with independent t-test). Data represent the mean ± SD of triplicate determinations from three independent experiments. CCNA2, cyclin A2; NEK2, NIMA related kinase 2; WHO, World Health Organization; TCGA, The Cancer Genome Atlas; GBM, glioblastoma; CGGA, Chinese Glioma Genome Atlas.