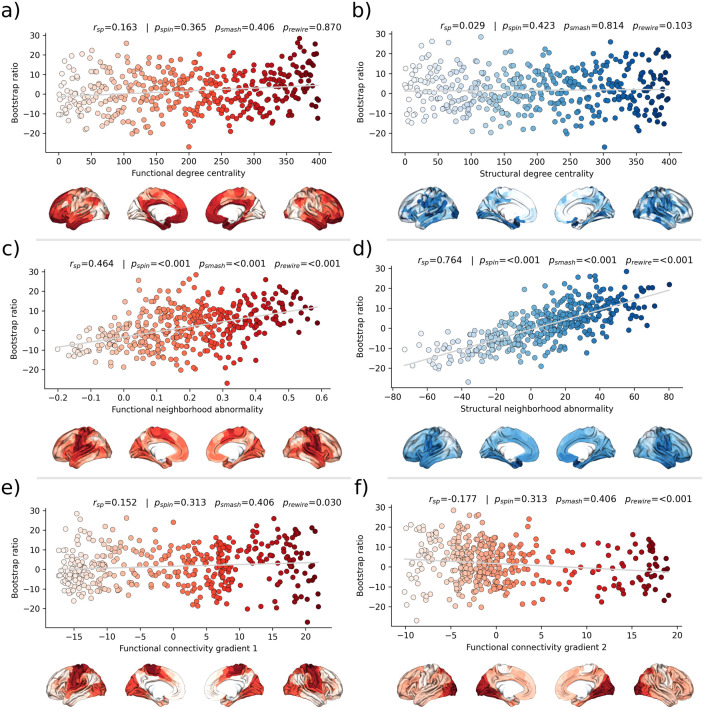
Figure 5. Brain network contextualization.
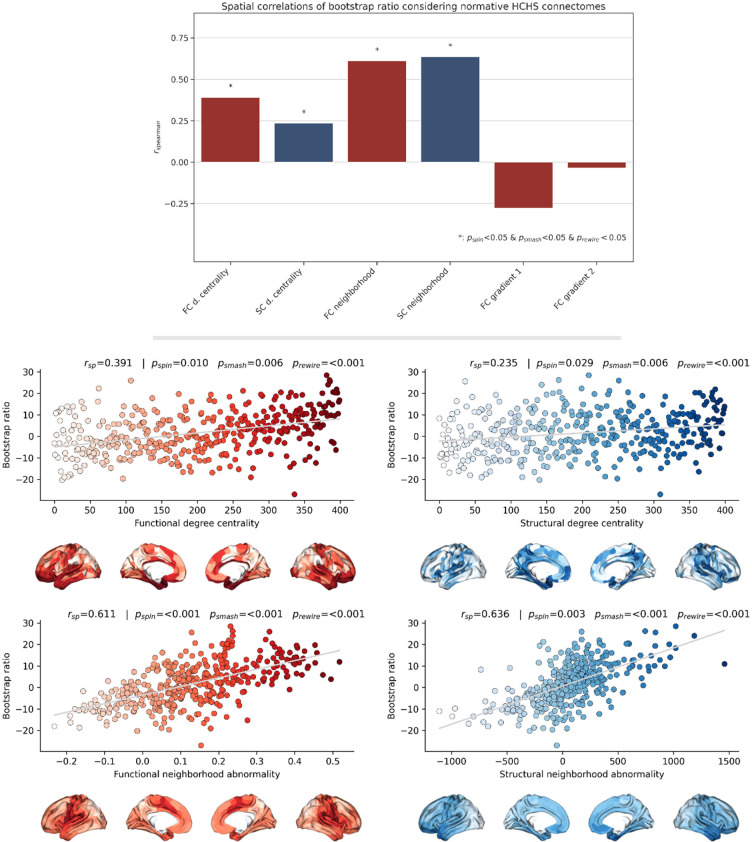
Spatial correlation results derived from relating Schaefer 400×7-parcellated maps of metabolic syndrome (MetS) effects (bootstrap ratio) to network topological indices (red: functional connectivity, blue: structural connectivity). Scatter plots that illustrate the spatial relationship are supplemented by surface plots for anatomical localization. The color coding of cortical regions and associated dots corresponds. (a and b) Functional and structural degree centrality rank. (c and d) Functional and structural neighborhood abnormality. (e and f) Intrinsic functional network hierarchy represented by functional connectivity gradients 1 and 2. Complementary results concerning t-statistic maps derived from group comparisons between MetS subjects and controls are presented in Figure 5—figure supplement 1. Corresponding results after reperforming the analysis with HCHS-derived group-consensus connectomes are presented in Figure 5—figure supplement 2. Abbreviations: HCHS – Hamburg City Health Study; - p-value derived from network rewiring (Maslov et al., 2004); - p-value derived from brainSMASH surrogates (Burt et al., 2020); - p-value derived from spin permutation results (Alexander-Bloch et al., 2018); - Spearman correlation coefficient.
Figure 5—figure supplement 1. Sensitivity network contextualization analysis based on t-statistic map derived from group comparison.