FIGURE 5.
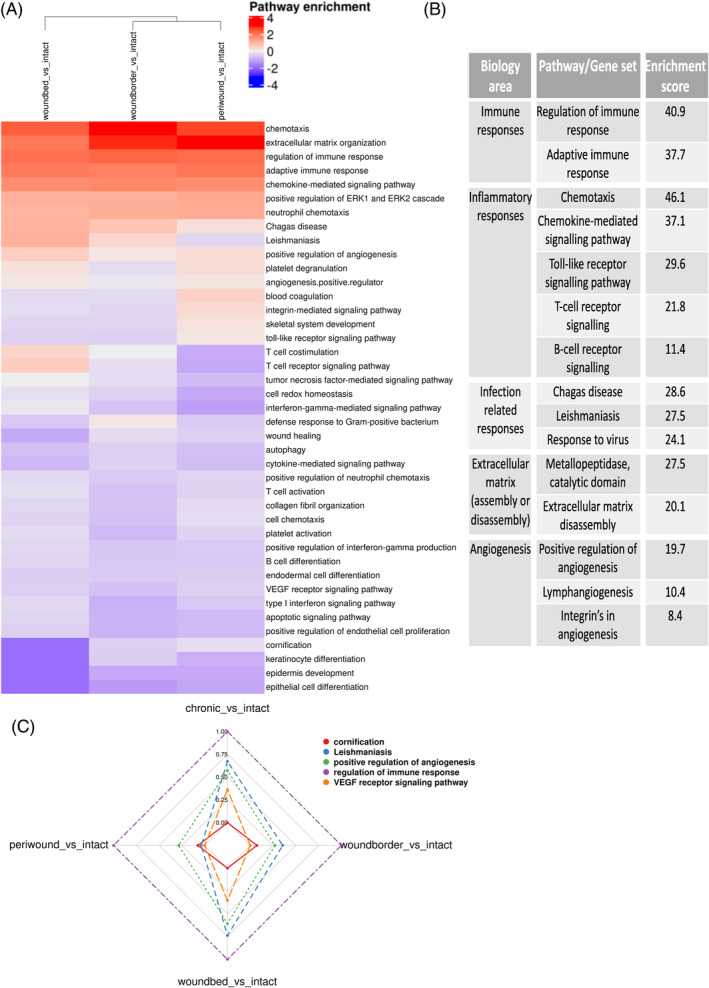
Molecular pathways and processes dysregulated in human chronic wounds and WHCs. (A) Heatmap clustering of pairwise comparisons of up‐ and downregulated pathways enriched in each wound subregion versus intact skin. Gene set enrichment analysis (GSEA) was performed using Biosystem, Interpro, and Gene Ontology sets, and only upregulated genes compared against intact skin were included for further analysis. The rows correspond to enriched pathways, and columns correspond to comparisons of wound subregions with intact skin. The column dendrogram is calculated using the Euclidean distance metric generated from the enrichment results for each comparison. The scaled, log10 converted enrichment results are colour‐coded (blue: downregulated enriched, white: not regulated, red: upregulated enriched; see the legend, top right). (B) Summary of some of the dysregulated pathways with enrichment results, grouped by biological area. (C) Radar plot displaying a selection of biological processes/pathways enriched across the different wound regions compared to intact skin samples. The selected pathways (color‐coded, top right) include cornification, Leishmaniasis, positive regulation of angiogenesis, regulation of immune response and VEGF receptor signalling pathway, representing the various processes dysregulated in chronic wounds and WHCs.
