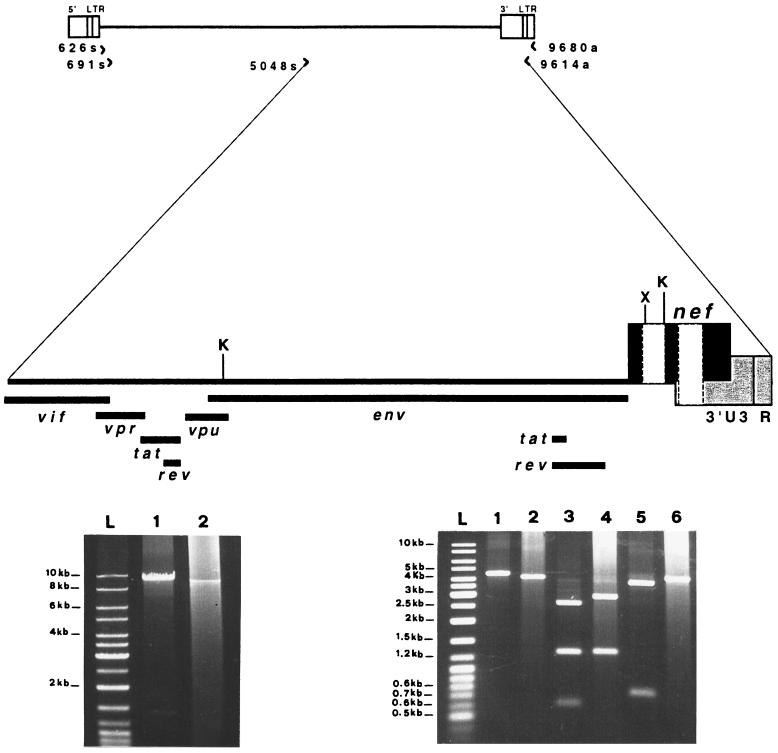
FIG. 7.
LD-PCR analysis of nearly full-length HIV-1 provirus (about 9 kb) and restriction enzyme analysis of the 3′ half of the viral genome (about 4.5 kb). (Top) Schematic illustration of HIV-1 proviral DNA showing the positions and the orientations of the nested PCR primers used to generate either the 9- or 4.5-kb fragment. The magnified 3′ half of the provirus shows the locations of the regulatory and accessory genes, the restriction sites (K, KpnI; X, XhoI), and two nef deletions (white boxes). The reference is the HIV-1 NL4-3 map. (Bottom left) Agarose gel electrophoresis (0.8%) of the second-round PCR products obtained from the 9-kb amplification (nested primer pairs, 626s-9680a for first round and 691s-9614a for second round). Lane 1, pNL4-3 (10 fg of plasmid diluted in DNA extracted from PBMC of a seronegative subject). Lane 2, SG1 (DNA extracted from 2 × 105 PBMC). Lane L, molecular size ladder (MBI, Fermentas). (Bottom right) Restriction site analysis of the 3′ half of the viral genome amplified by the nested primer pairs 626s-9680a (first round) and 5048s-9614a (second round). Lane 1, undigested pNL4-3; lane 2, undigested SG1; lanes 3 and 5, pNL4-3 digested with KpnI and XhoI, respectively; lanes 4 and 6, SG1 digested with KpnI and XhoI, respectively.