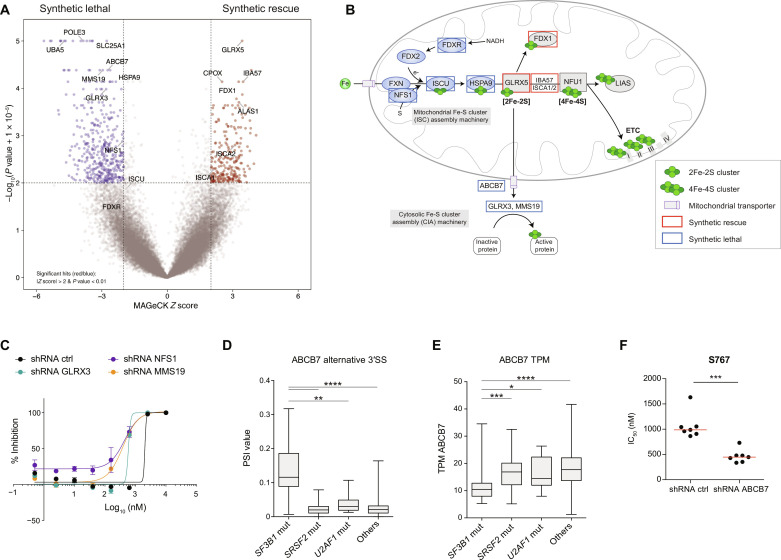
Fig. 3. CRISPR-Cas9 screen identifies ABCB7 as a sensitizing gene to S767-mediated cell death.
(A) Volcano plot representing the results of whole-genome CRISPR-Cas9 loss-of-function screen performed in EKO OCI-AML5 cells upon exposure to the S767 compound (1.1 μM). (B) Simplified representation of the ISC biosynthetic pathways. Briefly, ISCs are assembled in the mitochondrial matrix by the ISC assembly machinery on the scaffold protein ISCU. Clusters are transferred to ISC trafficking proteins through HSPA9 to be loaded on targeted mitochondrial proteins or transferred to form 4Fe-4S clusters. NFU1 lastly transfers 4Fe-4S clusters to target proteins including ETCI-III and LIAS, among others. ISC are also exported to the cytoplasm, putatively through the ABCB7 transporter, and loaded to cytosolic and nuclear proteins by the CIA machinery, comprising GLRX3 and MMS19 proteins, among others. Top hits from the Fe-S cluster pathway identified as synthetic lethal and synthetic rescue genes in the CRISPR-Cas9 screen are framed in blue and red, respectively. (C) Representative dose-response curves for S767 obtained in OCI-AML5 cells constitutively expressing shRNAs targeting key genes of the ISC biogenesis (error bars indicate SD of technical duplicates). (D) Box plot representation of the percent-splice-in (PSI) value regarding ABCB7 alternative 3′ splice site in primary AML specimens with or without mutations in the splicing factors SF3B1, SRSF2, or U2AF1 (Mann-Whitney U test). (E) ABCB7 mRNA expression expressed in TPM (transcripts per million) in primary AML samples carrying mutations in the indicated splicing factors (Mann-Whitney U test). (F) Dot plot distribution of S767 IC50 values in OCI-AML5 cells expressing an shRNA control or targeting ABCB7. Median is represented in red (n = 7, Mann-Whitney U test).